Figure 5.
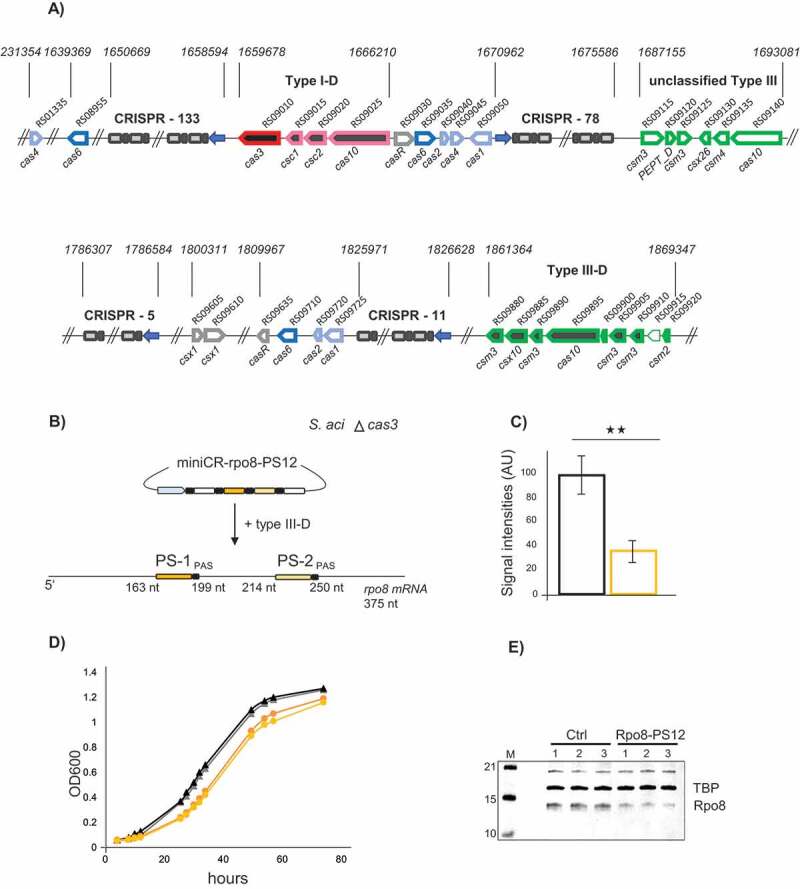
Silencing of rpo8 in S. acidocaldarius using the native type III-D system. A) Schematic representation of the CRISPR-Cas loci in S. acidocaldarius including all characterized adaptation cassettes (light blue), cas6 processing genes (blue), csx1 and casR accessory genes (grey), type I and type III effector complexes (pink, green) as well as CRISPR arrays (spacer number indicated); the unclassified type III cluster is of unknown function [21]; pseudogenes without annotation are indicated by a light border; the gene cas3 (red) was deleted in the mutant strain used in the rpo8 silencing experiment. B) Schematic representation of the silencing experiment using miniCR-rpo8-PS12 targeting the rpo8 mRNA at two protospacers (PS1, PS2); PS positions in respect to the gene length are indicated. C) Relative expression of the rpo8 mRNA relative to the 16S rRNA measured by RT-qPCR on reverse transcribed RNA extracts of control (black left bar) and rpo8 silenced transformants (orange right bar), harvested at OD600 = 0.3 each. Error bars, mean ± SD (n ≥ 3); Significant differences to Ctrl are indicated by asterisks (two-tailed t test, n ≥ 3, p ≤ 0.01). D) Growth profiles (OD600) of control (black triangles) and rpo8 silenced cultures (orange circles); two representative replicates grown up from separate colonies are shown. E) Western blot analyses using Anti-Rpo8 detecting Rpo8 (lower band) and Anti-TBP binding the transcription factor TBP (loading control, upper band). Three biological replicates of control (left) and rpo8- silenced cultures (harvested at OD600 = 0.3) are presented
