Figure 3.
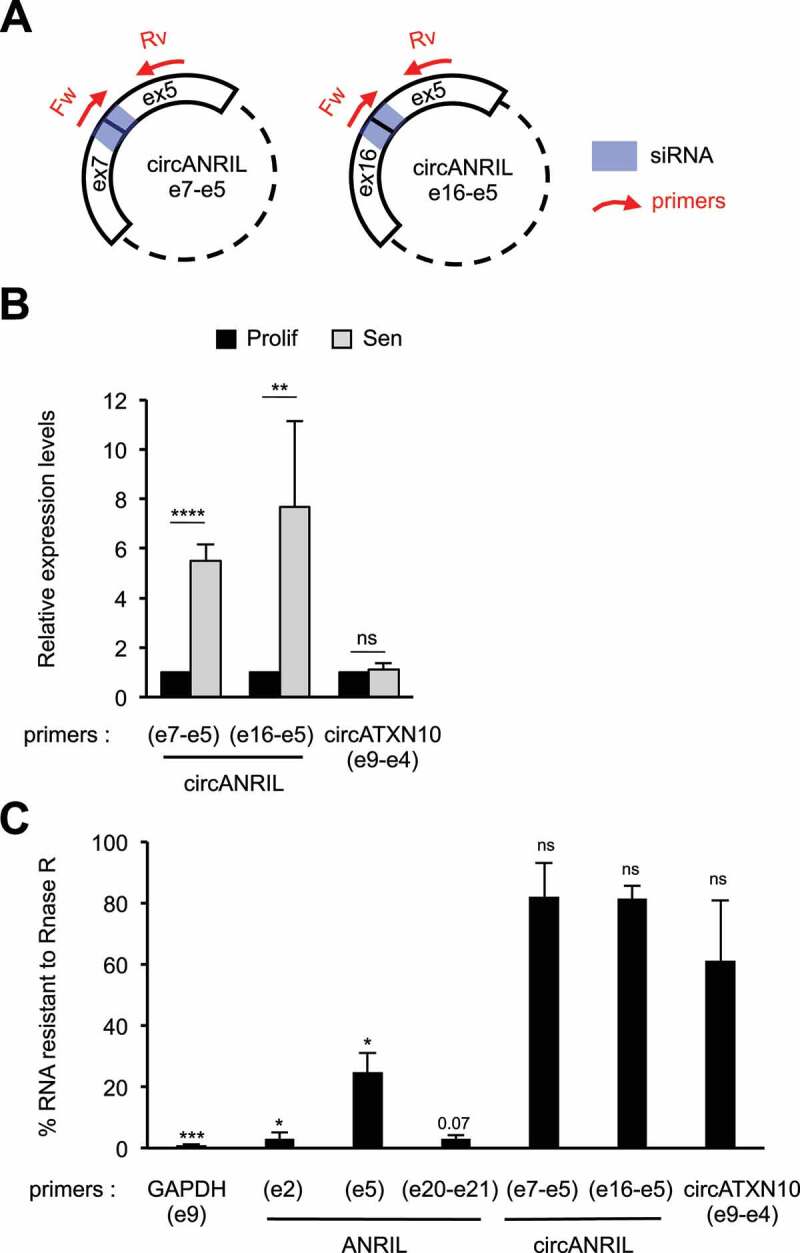
Circular ANRIL species are strongly induced in senescent cells
(A) Schematization of the circular ANRIL isoforms containing the specific back-spliced junctions that we identified in our study (Fig. S6). The primers and siRNAs targeting these back-spliced junctions are shown in red and blue, respectively. Note that although these primers give a unique PCR product, they can detect all circular species containing these specific back-spliced junctions, irrespective of the number or identity of spliced exons or introns located between exon 5 and the reversed spliced exon. (B) Total RNA was extracted from proliferative or senescent WI38 hTERT RAF1-ER cells. The expression of circular ANRIL species was measured by RT-qPCR using the indicated primers. CircATXN10 was measured as a control whose expression does not change in RAF1-induced senescence. The levels of RNA were normalized to those of GAPDH (e9) and then normalized to 1 in proliferative cells for each experiment. The means and standard deviations from four independent experiments are shown. Significant differences are indicated with asterisks (*: p value < 0.05, ** to ****: p values < 10−2, 10−3 and 10−4, respectively; two-sided paired Student’s t-test on log2 values), the number of the p value is indicated when it is between 0.05 and 0.1; ns: not significant. (C) Total RNAs from senescent WI38 hTERT RAF1-ER cells were treated or not with RNase R to digest all linear RNAs, and analysed by RT-qPCR. Percentages of RNAs resistant to RNase R treatment were calculated relative to the levels of these RNAs measured in untreated samples. Primers designed for e20-e21 junction detect both spliced and unspliced products, the intron 20 being very small (93 nt). Means and standard deviations from three independent experiments are shown (only two for ANRIL e20-e21, circANRIL e7-e5 and circANRIL e16-e5 primers). Significant differences are indicated as in (B).
