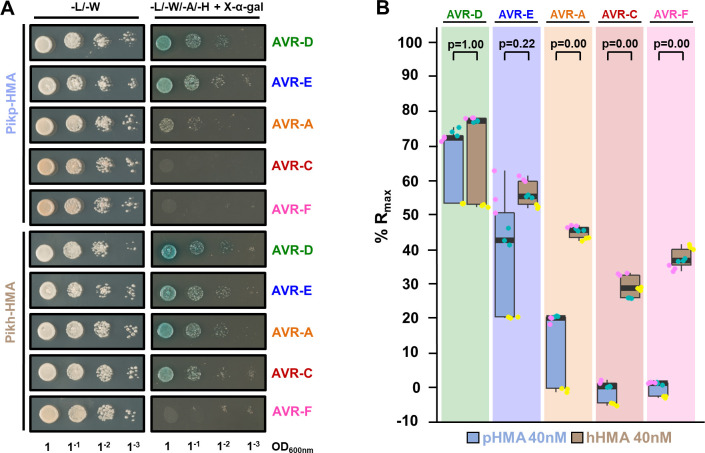
Fig 2. Pikh-HMA has increased binding to AVR-Pik effector alleles in vivo and in vitro.
(A) Yeast two-hybrid assay of Pikp-HMA and Pikh-HMA with AVR-Pik variants. For each combination of HMA/AVR-Pik, 5μl of yeast were spotted and incubated for ~60 h in double dropout plate for yeast growth control (left) and quadruple dropout media supplemented with X-α-gal (right). Growth, and development of blue colouration, in the selection plate are both indicative of protein:protein interaction. HMA domains were fused to the GAL4 DNA binding domain, and AVR-Pik alleles to the GAL4 activator domain. Each experiment was repeated a minimum of three times, with similar results. (B) Measurement of Pikp-HMA and Pikh-HMA binding to AVR-Pik effector variants by surface plasmon resonance. The binding is expressed as %Rmax at an HMA concentration of 40 nM. Pikp-HMA and Pikh-HMA are represented by blue and brown boxes, respectively. For each experiment, three biological replicates with three internal repeats each were performed, and the data are presented as box plots. The centre line represents the median, the box limits are the upper and lower quartiles, the whiskers extend to the largest value within Q1-1.5× the interquartile range (IQR) and the smallest value within Q3 + 1.5× IQR. All the data points are represented as dots with distinct colours for each biological replicate. “p” is the p-value obtained from statistical analysis and Tukey’s HSD. For results of experiments with 4 and 100 nM HMA protein concentrations, see S5 Fig.