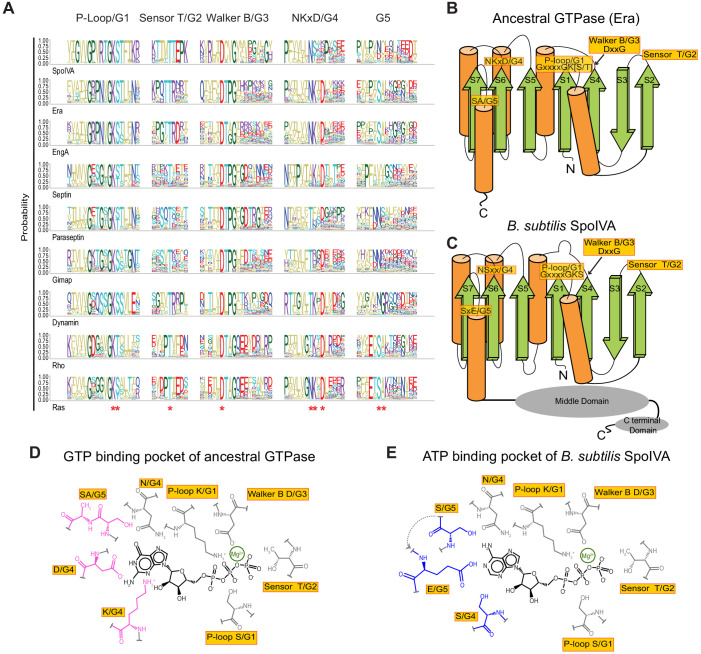
Figure 1. Predicted residues in SpoIVA evolved from an ancestral GTPase to bind ATP.
(A) Sequence logo displaying conservation of amino acid residues in different members of the TRAFAC class of GTPases. Letters represent amino acid abbreviations; height of each letter represents the probability of conservation among orthologs of the indicated protein. Red asterisk below the sequence logo indicates absolute conservation of the amino acid at that position. (B and C) Topological representation of (B) ancestral TRAFAC GTPase or (C) SpoIVA. Motifs in the active site are indicated in yellow; numbering (G1–G5) corresponds to an idealized GTPase (Bourne et al., 1991). N: amino terminus; C: carboxy terminus. β-strands are depicted as green arrows; α-helices are depicted as orange cylinders. Middle and C-terminal domains of SpoIVA are depicted as gray ovals. (D and E) Depiction of the nucleotide-binding pocket of (D) ancestral TRAFAC GTPase bound to GTP or (E) SpoIVA bound to ATP. Residues in the ancestral GTPase that contact the guanine base of GTP are depicted in pink; predicted residues in SpoIVA that may bind the adenine base of ATP are depicted in blue.