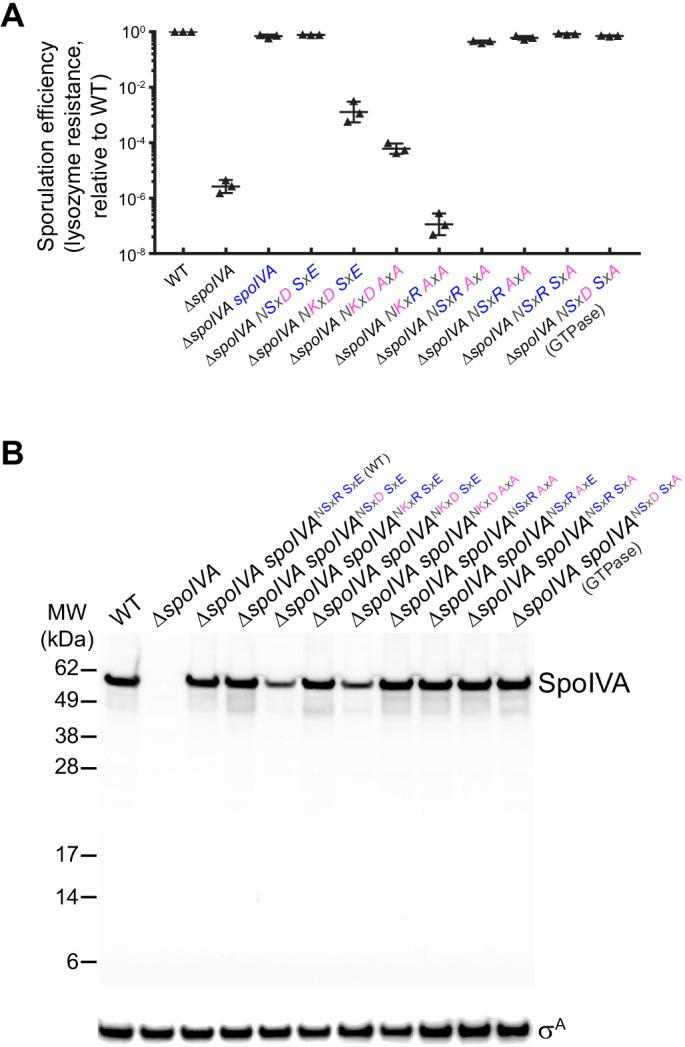
Figure 3. SpoIVAGTPase variant is functional in vivo.
(A) Sporulation efficiencies, relative to WT (PY79) and measured as resistance to 80°C for 20 min, of Bacillus subtilis strains (PY79, KP73, KR394, NG7, NG13, NG8, TU209, TU211, TU212, TU213, and TU223) harboring the indicated allele of spoIVA. Data points represent sporulation efficiencies from independent cultures (n = 3–4); bars indicate mean values; error bars are S.D.; ‘<10−8’ indicates that no heat-resistant spores were recovered. Sporulation efficiencies are listed in Supplementary file 2. (B–J) Fluorescence micrographs of sporulating B. subtilis strains (SL55, JH19, JH20, JH21, TU200, TU201, TU202, TU203, and TU227) harboring the indicated SpoIVA variant fused to green fluorescent protein imaged 3 hr after the onset of sporulation. (B–J) Fluorescence from GFP; (B’–J’) overlay, GFP fluorescence from B to J , respectively, and fluorescence from membranes visualized using FM4-64. Genotypes are listed in Supplementary file 1.
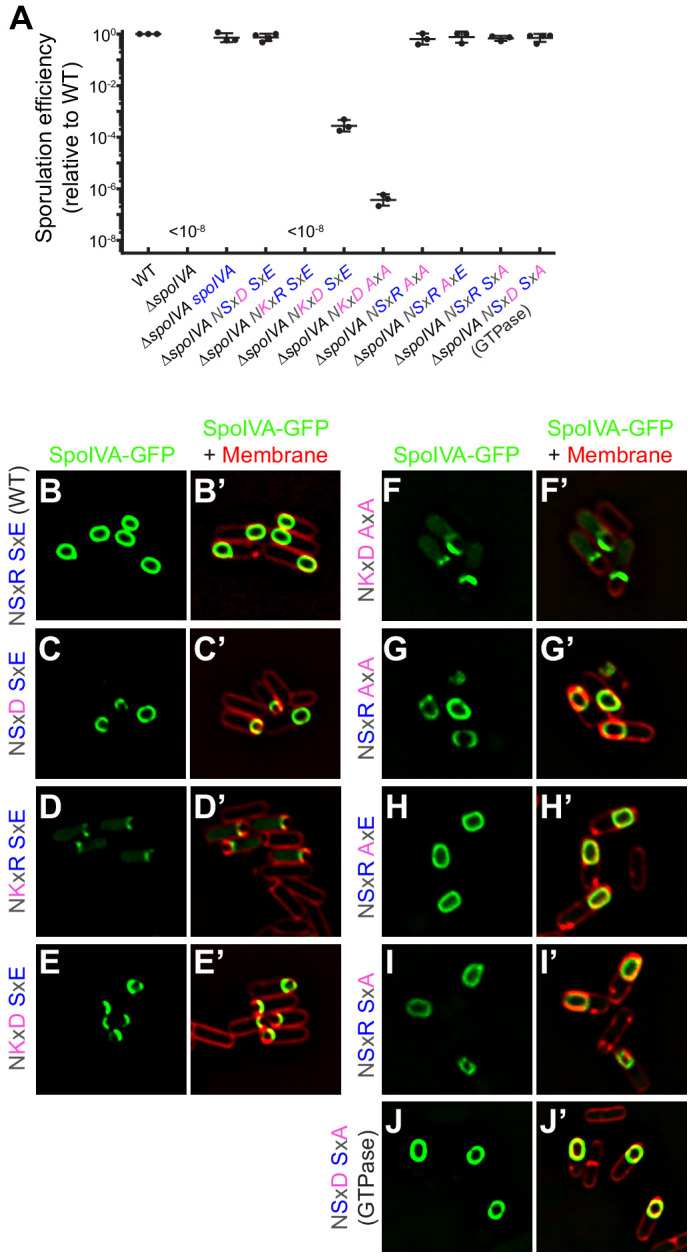
Figure 3—figure supplement 1. Lysozyme resistance and intracellular accumulation of SpoIVA and SpoIVA variants.