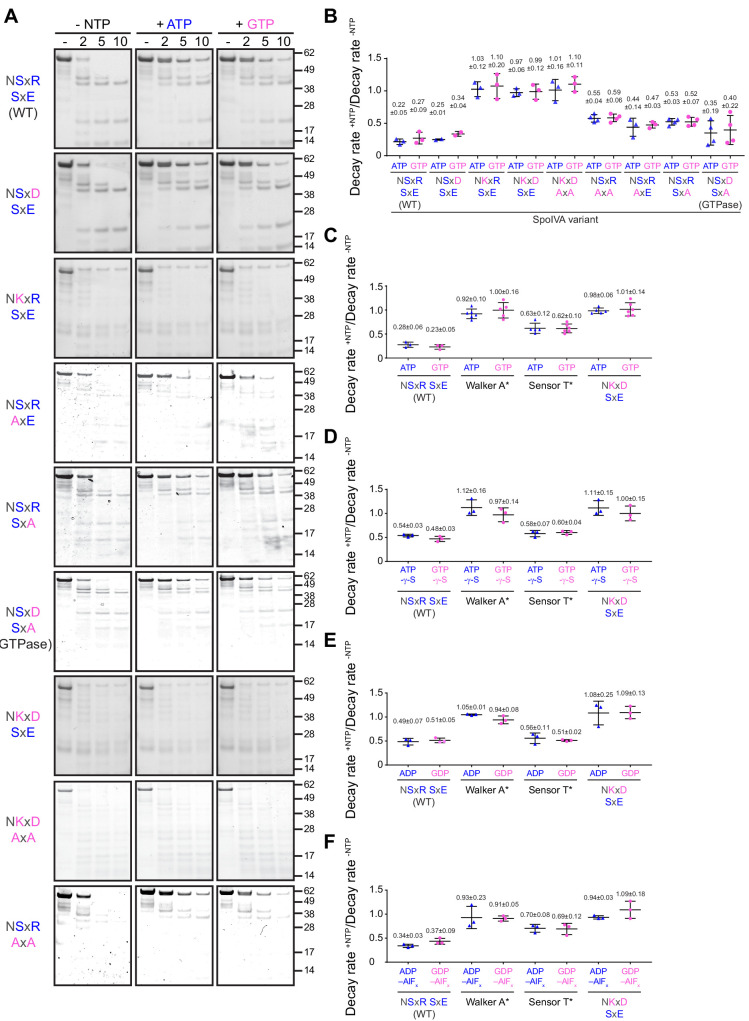
Figure 4. ATP or GTP hydrolysis, but not ADP or GDP binding, drives a conformational change in SpoIVA required for polymerization.
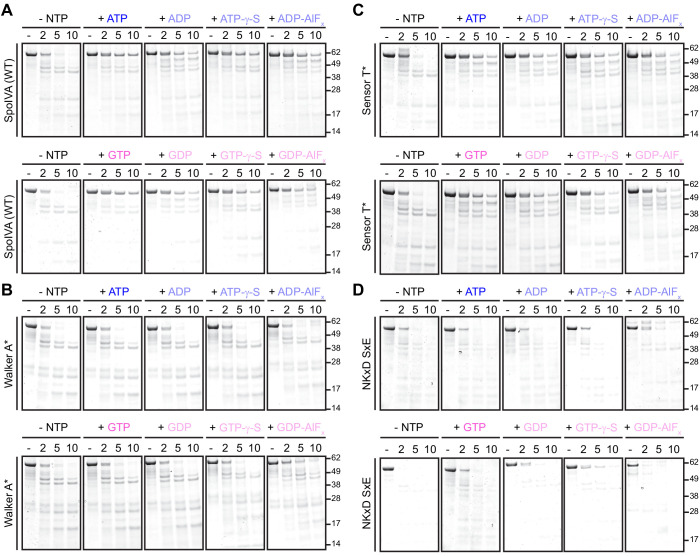
(A) Purified variants of SpoIVA at 2 µM (below the threshold concentration for polymerization) were incubated either in the absence of nucleotide (left panels) or in the presence of ATP (middle) or GTP (right) at 37°C for 4 hr. Reactions were then exposed to limited proteolysis by trypsin for the indicated times (2, 5, or 10 min), after which proteolysis was stopped by addition of SDS sample buffer and the products were analyzed by Coomassie-stained PAGE. Mobility of molecular weight markers (kilodaltons) are indicated to the right. Displayed is a representative image (n = 3–4) (B) Quantification of the disappearance of the full length purified SpoIVA variants in (A) in the presence of ATP (blue triangles) or GTP (pink circles). Rates of decay are reported as a ratio of that in the presence to the absence of nucleotide (Supplementary file 2). (C–F) Quantification of the disappearance of the full length purified SpoIVA variant indicated (WT; Walker A* which does not bind ATP; Sensor T* which binds but does not hydrolyze ATP; NKxD SxE which hydrolyzes ATP at an increased rate) as in (B) in the presence of (C) ATP or GTP; (D) ATP-γ-S or GTP-γ-S; (E) ADP or GDP; or (F) ADP-AlFx or GDP-AlFx. Representative images of Coomassie-stained gels for (C–F) are in Figure 4—figure supplement 1. Data points represent decay rate ratios from independent assays (n = 3–4); bars indicate mean values; error bars are S.D.