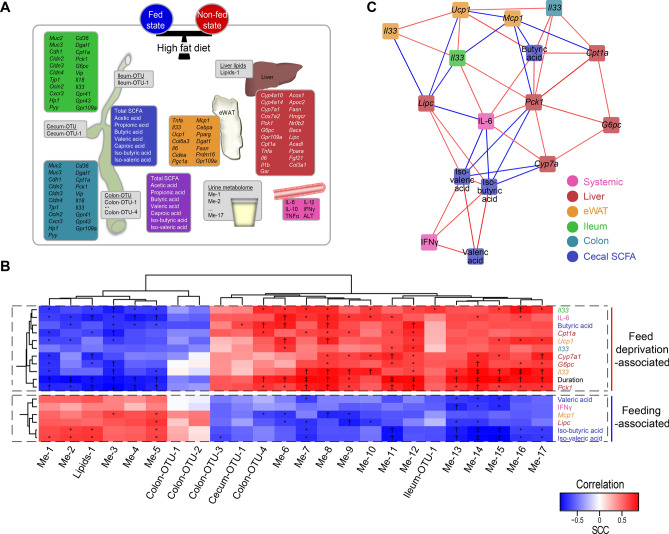
Figure 1.
Integration of host parameters and bacterial fermentation products with feed deprivation-induced gut microbiota, liver lipids, and urine metabolite clusters. (A) Metabolic and inflammatory host response parameters, gut microbiota, and intestinal fermentation products determined in fed (n = 10) and feed deprived (n = 9) high-fat diet-induced obese mice. The coloring of boxes relates to measured specific factors: gene expression in liver (dark red), epididymal white adipose tissue (eWAT, orange), ileum (green) and colon (turquoise), plasma-derived cytokines and alanine aminotransferase (ALT) (pink), short-chain fatty acids from cecum (blue) and feces (purple). Feed deprivation-associated lipid species cluster, which is based on determined liver triglyceride and phospholipid species (Lipids-1), the 17 urine metabolite clusters (Me-1 to Me-17) and OTU clusters from the ileal, cecal and colonic compartments (Ileum-OTU-1, Cecum-OTU-1, Colon-OTU-1 through Colon-OTU-4) are shown in grey boxes. (B) Correlation heat map of the hierarchically clustered z-score-normalized low complexity parameters (y-axis) against the computed eigenvectors of lipid, metabolite and OTU clusters (x-axis) associated with feed deprivation. The duration of feed deprivation is included as a host parameter (span between 8 h and 16 min to 11 h and 32 min). The heat map color represents Spearman rank correlation coefficients (SCC), as visualized in the insert, and asterisks mark FDR-adjusted significant correlations (*, FDR < 0.05; †, FDR < 0.01; ‡, FDR < 0.001). (C) Network showing the internal relationship among host parameters associated with feed deprivation for edges with significant SCC (red: SCC > 0.6, blue: rho < − 0.6, FDR < 0.05). Node colors indicate the tissue origin of the individual variable, as indicated in the legend.