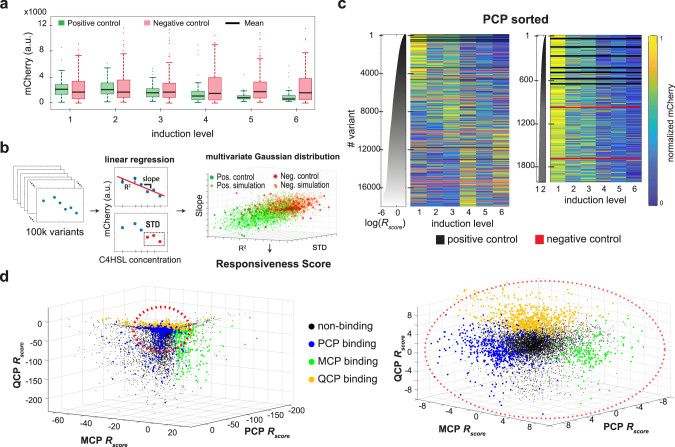
Fig. 2. Responsiveness analysis and results.
a Boxplots of mCherry levels for the positive (green) and negative (red) control variants for each of the six induction levels for PCP-GFP, based on 75 different variants for positive control and 187 different variants for negative control. On each box, the central mark indicates the median, and the bottom and top edges of the box indicate the 25th and 75th percentiles, respectively. The value for ‘Whisker’ corresponds to approximately ±2.7 STD (standard deviation) and 99.3 percent coverage and extends to the adjacent value, which is the most extreme data value that is not an outlier. The outliers are plotted individually as plus signs. b Schema for responsiveness score (Rscore) analysis. (Left and middle) Linear regression was conducted for each of the 100k variants, and two parameters were extracted: slope and goodness of fit (R2). The third parameter is the standard deviation (STD) of the fluorescence values at the three highest induction levels. (Right) Location of the positive control (dark green stars) and negative control (red stars) in the 3D-space spanned by the three parameters. Both populations (positive and negative) were fitted to 3D-Gaussians, and simulated data points were sampled from their probability density functions (pdfs) (orange for negative and green for positive). Based on these pdfs the Rscore was calculated. c (Left) Heatmap of normalized mCherry expression for the ~20k variants with PCP. Variants are sorted by Rscore. Black and red lines are positive and negative controls, respectively, and the gray graph is the Rscore as a function of variant. (Right) “Zoom-in” on the 2,000 top-Rscore binding sites for PCP. d (Left) 3D-representation of the Rscore for every binding site in the library and all RBPs. Responsive binding sites, i.e., sites with Rscore > 3.5, are colored blue for PCP, green for MCP, and yellow for QCP. (Right) “Zoom-in” on the central highly concentrated region. Source data are provided as a Source data file. Altogether, we identified 1868, 1144, and 2624 binding sites (i.e., Rscore > 3.5) for PCP, MCP, and QCP, respectively. In addition, there were an additional 3736, 1460, and 4682 “non-classified” binding sites (i.e., 0 < Rscore < 3.5) for PCP, MCP, and QCP, respectively, while the rest were determined to be non-binding (Rscore < 0).