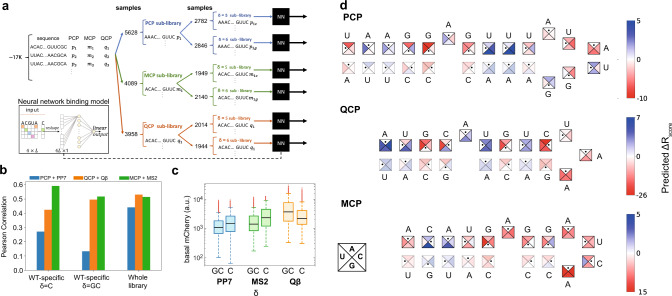
Fig. 3. Analysis of MCP, PCP, and QCP RNA-binding sequence preferences.
a Scheme for the data preparation and neural network architecture (inset) used. b Pearson correlation over a held-out test set computed for the WT-specific sub-libraries (i.e., PCP, MCP, and QCP with PP7-based, MS2-based, and Qβ-based binding sites, respectively at either δ = C (left) or δ = GC (middle)), and for the whole-library CNN model (right). c Boxplots of basal mCherry levels for the six WT-specific sub-libraries, based on the following number of strains (from left to right): 3113 (PP7-GC), 3067 (PP7-C), 2810 (MS2-GC), 2743 (MS2-C), 2702 (Qβ-GC), 2743 (Qβ-C). On each box, the central mark indicates the median, and the bottom and top edges of the box indicate the 25th and 75th percentiles, respectively. The value for ‘Whisker’ corresponds to approximately ±2.7 STD (standard deviation) and 99.3 percent coverage and extends to the adjacent value, which is the most extreme data value that is not an outlier. The outliers are plotted individually as plus signs. d Illustrations of the whole-library model predictions for the three sub-libraries for any single- or double-nucleotide structure-preserving mutation. Each binding site is shown, with the wild-type sequence indicated as black dots inside the squares. Each square is divided to the four possible options of nucleotide identity, with the colors representing the predicted change in Rscore with respect to the wild type for each option. Source data are provided as a Source data file.