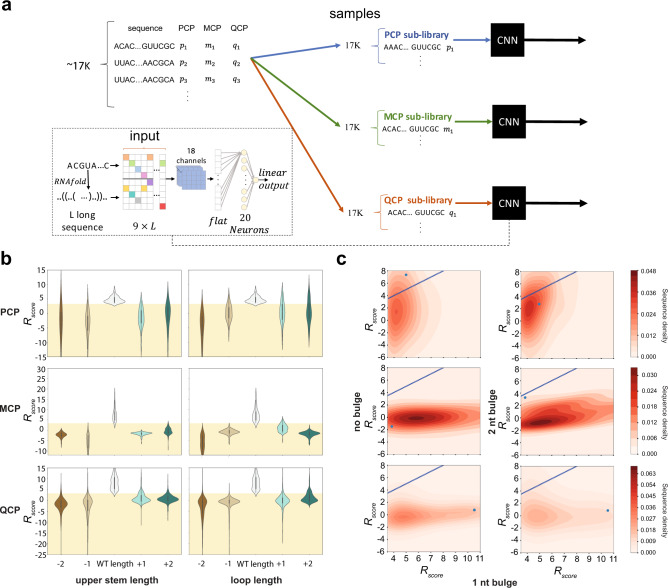
Fig. 4. Analysis of MCP, PCP, and QCP RNA-binding structure preferences.
a A scheme for the data preparation and neural network architecture (inset) used for the protein-specific CNN model based on the whole-library. We generated various binding sites with a predefined structure different from the wild type and used the whole-library models to predict their responsiveness score. b Predicted Rscore distributions for binding sites that differ in the length of the upper stem (left) or the loop (right) for PCP (top row), MCP (middle row), and QCP (bottom row). Stem and loop lengths were varied by ±2 base-pairs and nucleotides, respectively. Yellow shade corresponds to non-binding Rscore values. c Density maps for predicted Rscore for either no bulge (left-column) or a 2-nucleotide bulge (right-column) mutation of a wild-type-like structure for PCP-response (top row), MCP-response (middle row), and QCP-response (bottom row). Source data are provided as a Source data file. Blue dots correspond to wild-type sequences.