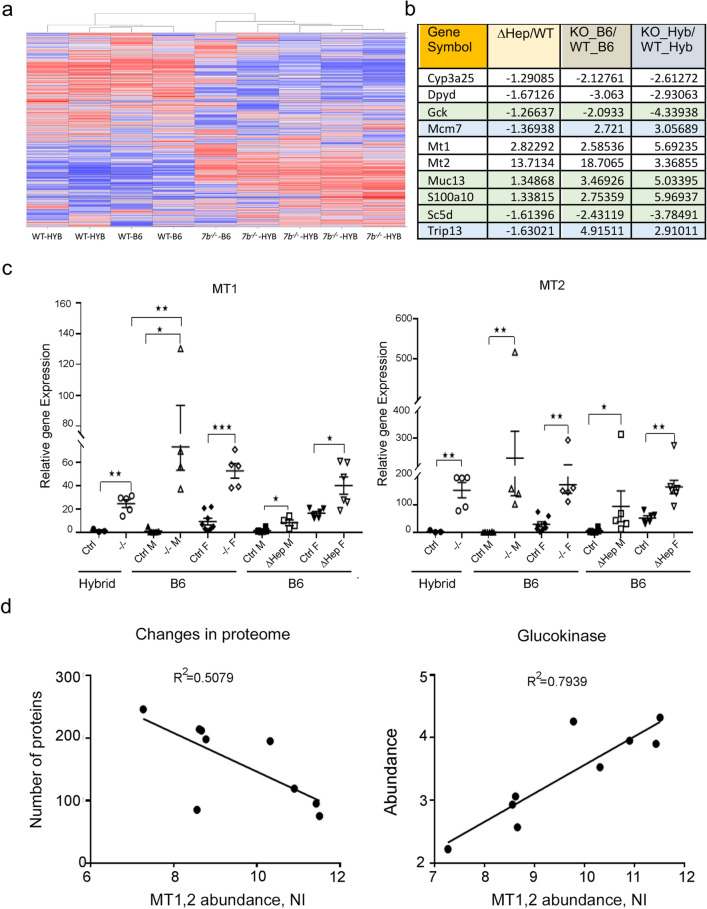
Figure 8.
Comparison of Atp7b mutant mice suggests potential markers and modifying factors. (a) Unsupervised clustering of protein profiles from the dataset # 2 (see methods) was done using Spotfire DecisionSite with Functional Genomics v9.1.2 (https://edelivery.tibco.com/storefront/eval/tibco-spotfire-decisionsite-for-functional-genomics/prod10196.html) and presented as a heat map; the map illustrates marked differences between the wt animals and Atp7b−/− global knockouts and presence of similarly changed proteins in Atp7b−/− mice on different genetic backgrounds heat map (b) The most significantly dysregulated proteins (|S|= 6, p value < 0.05) detected in all Atp7b-mutant mice. Green color highlights protein with changes in abundance that reflect the extent of liver pathology; blue indicate proteins that are oppositely changed in the hepatocyte-specific and global knockouts; (c) Upregulation of metallothioneins MT1 and MT2 in Atp7b-deficient strains. One-way ANOVA and a two-tail t-test for individual pairs (shown) were used (*p value ≤ 0.05, **p value ≤ 0.01, ***p value ≤ 0.001) and yielded same conclusions. “Ctrl” indicates either Atp7b+/+ or Atp7b +/- animals; “M”—males; “F”—females. (d) Left: Correlation between the abundance of metallothioneins MT1 and MT2 in individual Atp7b−/− animals and the number of proteins in their liver proteome changed more than1.5-fold compared to the averaged wild-type on the same genetic background. Right: Correlation between abundances of MT1 and MT2 and levels of glucokinase in individual Atp7b−/− animals. “NI” indicates a normalized protein intensity.