Abstract
The post-transcriptional regulator RsmA globally controls gene expression in bacteria. Previous studies showed that RsmA2 and RsmA3 played critical roles in regulating type III secretion system (T3SS), motility, syringafactin, and alginate productions in Pseudomonas syringae pv. tomato strain DC3000 (PstDC3000). In this study, we investigated global gene expression profiles of the wild-type PstDC3000, the rsmA3 mutant, and the rsmA2/A3 double mutant in the hrp-inducing minimum medium (HMM) and King’s B (KB) medium. By comparing the rsmA2/A3 and rsmA3 mutants to PstDC3000, a total of 1358 and 1074 differentially expressed genes (DEGs) in HMM, and 870 and 1463 DEGs in KB were uncovered, respectively. When comparing the rsmA2/A3 mutant with the rsmA3 mutant, 277 and 741 DEGs in HMM and KB, respectively, were revealed. Transcriptomic analysis revealed that the rsmY, rsmZ, and rsmX1-5 non-coding small RNAs (ncsRNAs) were positively affected by RsmA2 and RsmA3, while RsmA3 positively regulates the expression of the rsmA2 gene and negatively regulates both rsmA1 and rsmA5 gene expression. Comparative transcriptomic analysis showed that RsmA2 and RsmA3 synergistically influenced the expression of genes involved in T3SS and alginate biosynthesis in HMM and chemotaxis in KB. RsmA2 and RsmA3 inversely affected genes involved in syringafactin production in HMM and ribosomal protein biosynthesis in KB. In addition, RsmA2 played a major role in influencing genes involved in sarcosine and thiamine biosynthesis in HMM and in mannitol and phosphate metabolism in KB. On the other hand, genes involved in fatty acid metabolism, cellulose biosynthesis, signal transduction, and stress responses were mainly impacted by RsmA3 in both HMM and KB; whereas RsmA3 played a major role in controlling genes involved in c-di-GMP, phosphate metabolism, chemotaxis, and capsular polysaccharide in HMM. Furthermore, regulation of syringafactin production and oxidative stress by RsmA2 and RsmA3 was experimentally verified. Our results suggested the potential interplay among the RsmA proteins, which exhibit distinct and overlapping roles in modulating virulence and survival in P. syringae under different nutritional conditions.
Keywords: Pseudomonas syringae, RsmA, CsrA, non-coding small RNA, T3SS, syringafactin, ROS
Introduction
Pseudomonas syringae pv. tomato strain DC3000 (PstDC3000), a pathogen of tomato, Brassica spp. (cabbage and cauliflower), and Arabidopsis thaliana (Whalen et al., 1991; Wang et al., 2002; Sreedharan et al., 2006), is a model strain for studies in molecular mechanisms of bacterial pathogenesis and in plant–microbe interactions (Xin and He, 2013). Over 30 effectors in PstDC3000 have been identified to be secreted and translocated into host cells via the type III secretion system (T3SS) to promote disease (Boch et al., 2002; Guttman et al., 2002; Zhao et al., 2003). The expression of the T3SS genes is activated by a HrpL-RpoN sigma factor cascade and bacterial enhancer-binding proteins (EBPs) HrpRS (Alarcón-Chaidez et al., 2003; Tang et al., 2006; Xie et al., 2019). Besides T3SS, phytotoxin coronatine (COR), extracellular protease, and alginate all contribute to the virulence of PstDC3000 (Brooks et al., 2004; Ishiga et al., 2018).
Previous studies have reported that the GacS/GacA two-component system (TCS) affects virulence via regulation of motility, biofilm formation, quorum sensing (QS), stress response, secondary metabolites, and production of extracellular enzymes (Heeb and Haas, 2001; Lapouge et al., 2008; Sonnleitner et al., 2009). GacS, a sensor kinase, senses one or more signals and phosphorylates itself and GacA, a response regulator (Heeb and Haas, 2001). Phosphorylated GacA specifically activates non-coding small RNAs (ncsRNAs), e.g., csrB and csrC, in Escherichia coli (Gudapaty et al., 2001; Suzuki et al., 2002), rsmY and rsmZ in Pseudomonas aeruginosa (Kay et al., 2006; Janssen et al., 2018), and rsmX1-5, rsmY, and rsmZ in PstDC3000 (Moll et al., 2010; Ge et al., 2019). The ncsRNAs contain many GGA motifs which exhibit high affinity with the RNA-binding protein CsrA (carbon storage regulator) or its homologs RsmA and RsmE (repressor of secondary metabolites), thus sequestering and antagonizing their functions (Reimmann et al., 2005; Duss et al., 2014; Vakulskas et al., 2015).
As post-transcriptional regulators, the CsrA/RsmA family proteins bind to specific GGA motifs of the RNA secondary structures in the 5′ untranslated regions (UTRs), thus affecting mRNA stability, riboswitch function, transcript elongation, and repressing or activating translation of target genes (Schubert et al., 2007; Vakulskas et al., 2015; Pourciau et al., 2020). It has been revealed that CsrA/RsmA proteins act as global virulence regulators for many bacterial pathogens in both animals and plants. The rsmA mutant of P. aeruginosa could not activate T3SS, resulting in reduced depolymerization, cytotoxicity, and anti-internalization during interaction with airway epithelial cells (Mulcahy et al., 2006). In Pseudomonas amygdali pv. phaseolicola, RsmA and RsmE redundantly controlled phaseolotoxin biosynthesis and virulence (Ramírez-Zapata et al., 2020). In Erwinia amylovora, CsrA positively regulated genes involved in T3SS, amylovoran production, and motility and activated the Rcs phosphorelay system by binding to rcsB (Ancona et al., 2016; Lee et al., 2019). In Xanthomonas citri, RsmA directly regulated the T3SS master regulator HrpG, and mutation of the rsmA gene decreased exopolysaccharide (EPS) production and abolished hypersensitive response (HR) in non-host plants (Andrade et al., 2014).
Pseudomonas syringae pv. tomato strain DC3000 contains five RsmA protein homologs, i.e., RsmA1 to RsmA5. Previous studies revealed that RsmA2 and RsmA3 were required for T3SS, motility, coronatine toxin, pyoverdine, syringafactin, and alginate productions and in planta development of disease symptoms and exhibited strong binding affinities to rsmX1, rsmX5, rsmY, and rsmZ ncsRNAs (Ge et al., 2019). In this study, we used RNA-seq to investigate the global gene expression profiles of PstDC3000 and the rsmA3 and rsmA2/A3 mutants in both the hrp-inducing minimum medium (HMM) and King’s B medium (KB). Comparative transcriptomic analysis revealed distinct and overlapping roles in gene regulation by RsmA2 and RsmA3 in PstDC3000.
Materials and Methods
Bacterial Strains and Growth Conditions
All strains used in this study were as reported previously (Ge et al., 2019). The wild-type PstDC3000 and its rsmA mutants were routinely cultured in KB medium at 28°C with shaking at 250 rpm. The HMM, supplemented with 10 mM fructose as carbon source, and KB medium were used for RNA isolation (Huynh et al., 1989; Ge et al., 2019). Bacterial growth was monitored by measuring the absorbance of cell suspensions at 600 nm. Antibiotics were supplied at the following final concentrations: 100 μg/ml rifampicin, 50 μg/ml kanamycin, and 100 μg/ml ampicillin.
RNA Extraction
Overnight cultures of the bacterial strains were collected by centrifugation and washed with HMM or KB for three times, respectively. The suspensions were adjusted to OD600 = 0.2 in HMM and KB and incubated at 18 and 28°C for 6 h, respectively. The OD values for samples were similar at collection time. Four ml of RNA protect reagent (Qiagen, Hilden, Germany) was added to 2 ml of bacterial culture mixed by vortex and incubated at room temperature for 5 min. Cells were harvested by centrifugation, and total RNAs were extracted using RNeasy® mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. DNase I treatment was performed with TURBO DNA-free kit (Ambion, Austin, TX, United States). The quantity and quality of RNA samples were determined using a Nano-drop ND100 spectrophotometer (Nano-Drop Technologies, Wilmington, DE, United States) and/or using Agilent RNA 6000 Nano Chip Bioanalyzer (Agilent, Santa Clara, CA, United States).
RNA-seq Analysis
Library construction and sequencing of three biological samples each of PstDC3000 and its rsmA mutants were performed using the Illumina HiSeq 4000 (Illumina, San Diego, CA, United States) by the Keck Center at the University of Illinois, Urbana-Champaign. Ribosomal RNA was removed with the Ribo-zero Bacteria kit (Illumina), and a total of 18 stranded libraries were constructed using the TruSeq Stranded RNA Sample Prep kit following the manufacturer’s instructions (Illumina, San Diego, CA, United States). The sequence reads were aligned to the genome of PstDC3000 (GenBank accession #: AE016853.1) (Buell et al., 2003) using Bowtie2 version 2.3.2 (Langmead and Salzberg, 2012). Samtools and bedtools were performed for getting the read counts per coding sequence (CDS). Normalized log2-based count per million values (log2CPM) was calculated after trimmed mean of M value (TMM) normalization using the edgeR package (Robinson et al., 2010).
To examine gene expression dynamics among all the samples, a multidimensional scaling (MDS) was drawn using glMDSPlot function in R. Differentially expressed genes (DEGs) were detected using edgeR and defined as genes with a | FC (Fold change)| value ≥ 1.5 and a corrected p value < 0.05 from three biological samples. To visualize the overall expression pattern of individual genes, the MA plots (a.k.a., mean-difference plots; log2FC versus average log2CPM; FC, fold change; CPM, counts per million reads) and heat maps were, respectively generated using plotMD and heatmap.2 functions in R. Protein sequences of all coding genes in PstDC3000 (gene bank accession #: AE016853.1) were downloaded from NCBI website1. The FASTA protein file was used as input for protein annotation using eggNOG-mapper2. Clusters of orthologous groups (COGs) information for DEGs was extracted from the eggNOG output file. In addition, genes involved in T3SS were manually grouped into an additional orthologous categorization. RNA-seq data files have been submitted to Gene Expression Omnibus (GEO) at the National Center for Biotechnology Information (NCBI) with an accession number GSE162091.
Identification of Genes Specifically Affected by RsmA2, RsmA3, or Both
Venn diagrams were drawn by analyzing the DEG lists for comparisons of the rsmA23 mutant versus PstDC3000, the rsmA23 mutant versus the rsmA3 mutant, and the rsmA3 mutant versus PstDC3000 in both HMM and KB. DEGs up-regulated (or down-regulated) simultaneously in all three comparisons were considered to be synergistically regulated by RsmA2 and RsmA3, whereas DEGs inversely expressed in two comparisons (rsmA23 versus rsmA3 and rsmA3 versus PstDC3000) were considered to be inversely regulated by RsmA2 and RsmA3. On the other hand, DEGs found in comparison of rsmA23 versus PstDC3000 and rsmA23 versus rsmA3, but not in rsmA3 versus PstDC3000 comparison, were considered to be mainly regulated by RsmA2, whereas DEGs found in comparison of rsmA23 versus PstDC3000 and ΔrsmA3 versus PstDC3000, but not in rsmA23 versus rsmA3 comparison, were considered to be mainly regulated by RsmA3.
Quantitative Real-Time PCR (qRT-PCR)
For quantitative Real-Time PCR (qRT-PCR), 1 μg of RNA was reversed to cDNA using the SuperScriptTM III Reverse Transcriptase following the manufacturer’s instructions (Invitrogen, Carlsbad, CA, United States). Concentration of cDNA was adjusted to 100 ng/μl and used as template for qRT-PCR. The PowerUp SYBR® Green PCR master mix (Applied Biosystems, Foster, CA, United States) was used to detect the gene expression of selected genes. The qRT-PCR amplifications were performed in the StepOnePlus Real-Time PCR system (Applied Biosystems, Foster, CA, United States) under the following procedure: 50°C for 2 min, and 95°C for 2 min followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. The rpoD was used as an endogenous control to calculate relative quantification (ΔΔCt) (Ge et al., 2019). All primers are listed in Supplementary Table 1. The experiment was repeated, and three biological replicates were used for each gene.
Detection of Syringafactin Production
Atomized oil assay was used to detect syringafactin as previously described (Burch et al., 2010). PstDC3000 and all rsmA mutants were grown on KB agar plates for 48 h, resuspended in PBS, and adjusted to an OD600 = 1.0. Ten microliters was pipetted onto the surface of KB plates (1.5% agar) and incubated for 24 h at 20°C. An airbrush (VIVO HOME, Pleasanton, CA, United States) was used to spray a mist of mineral oil over the plates (light paraffin oil; Thermo Fisher Scientific, Waltham, MA, United States). Brighter oil drops formed a visible halo around bacterial colonies. The ring area of the halos was measured to represent syringafactin production. Experiments were performed in triplicate and repeated three times. Statistical comparison among different strains was performed using one-way ANOVA followed by Fisher’s LSD test (p < 0.05).
Oxidative Stress Assay
Spot dilution assay was performed using a previously described procedure to detect oxidative sensitivity (Ge et al., 2018). Briefly, overnight bacterial cells were harvested by centrifugation and washed twice using PBS. After the final wash, the pellet was resuspended in PBS and adjusted to OD600 = 1. Tenfold serial dilutions of the bacterial suspension were made in PBS. Each dilution (5 μl) was spotted on the plates with different concentrations of H2O2 (0, 0.25, or 0.5 mM) and incubated at 28°C for 2 days. The experiment was performed in duplicate and repeated three times.
Results
Summary of Gene Expression Profiles
Previous studies showed that RsmA2 and RsmA3 played major roles in the virulence of PstDC3000 and the rsmA2/A3 double mutant exhibited dramatically reduced disease symptoms and in planta bacterial growth. Furthermore, RsmA2 and RsmA3 played distinct roles in regulating virulence factors, including T3SS and swarming motility (Ge et al., 2019). In order to further understand the global effects of RsmA2 and RsmA3, as well as their distinct roles in regulating gene expression, RNA-seq comparing the wild-type PstDC3000, the rsmA3 mutant (ΔrsmA3) and the rsmA2/A3 double mutant (ΔrsmA23) were performed in both HMM and KB media. In total, 11,352,295 to 13,869,462 reads for each biological sample were generated for PstDC3000, ΔrsmA3, and ΔrsmA23 grown in HMM, and the percentage of reads mapped to the PstDC3000 genome ranged from 81.2 to 99.7%, whereas 9,332,318 to 12,837,068 reads for each biological sample were obtained for PstDC3000, ΔrsmA3, and ΔrsmA23 in KB, and the percentage of reads mapped to PstDC3000 genome was from 60.3 to 98.5%.
To explore the similarities and differences between these samples, MDS was conducted. MDS plot clearly showed that the first two dimensions explained about 60 and 21% of the variability in the whole datasets, respectively (Supplementary Figure 1). Dimensions 1 and 2, respectively represented data variations due to different media and different strains (Supplementary Figure 1). The three biological samples of each treatment (strain/medium combination) were clustered together. Furthermore, the heat map also showed that the three biological samples of each strain in different media were very consistent (Supplementary Figure 2), indicating that the datasets were highly reproducible.
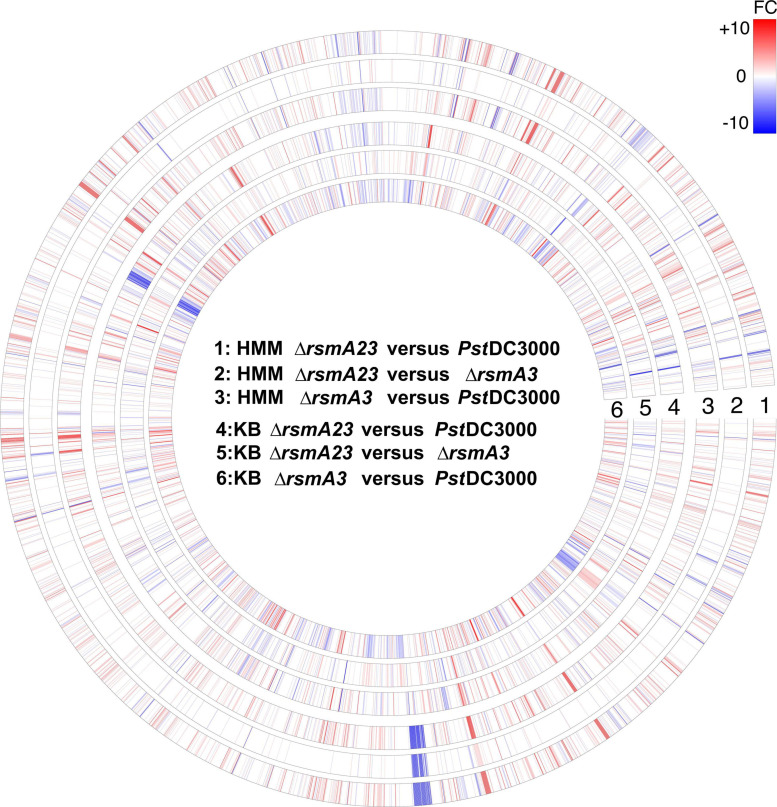
To show the overall transcriptomic profiles, a circle plot (Figure 1) was constructed where the expression of all 5348 genes was displayed by comparing ΔrsmA23 versus PstDC3000, ΔrsmA23 versus ΔrsmA3, and ΔrsmA3 versus PstDC3000 in both HMM and KB. Among the 5348 genes, a total of 2661 genes exhibited a differential expression with a | Fold change (FC)| value ≥ 1.5 and a p value < 0.05 between PstDC3000, ΔrsmA3, and ΔrsmA23 grown in both HMM and KB media (Supplementary Figure 2A). These 2661 genes were designated as DEGs, representing about half of all the genes in the PstDC3000 genome. Among the 2661 DEGs, 1560 and 1879 were differentially expressed in HMM and KB, respectively (Supplementary Figures 2B,C and Supplementary Tables 2, 3).
FIGURE 1.
Circular map of RNA-seq data for all 5348 genes. For each gene, red or blue color represents fold change (FC) ≥ 1.5 and FC ≤ –1.5 with a p value < 0.05, respectively. The higher the | FC|, the deeper the color. Rings # 1, 2, and 3 represent FCs for comparison of ΔrsmA23 versus PstDC3000, ΔrsmA23 versus ΔrsmA3, and ΔrsmA3 versus PstDC3000 in HMM, respectively. Rings # 4, 5, and 6 represent FCs for comparison of ΔrsmA23 versus PstDC3000, ΔrsmA23 versus rsmA3, and ΔrsmA3 versus PstDC3000 in KB, respectively. HMM: hrp-inducing minimum medium; KB: King’s B medium; PstDC3000: Pseudomonas syringae pv. tomato DC3000; ΔrsmA3: the rsmA3 mutant; ΔrsmA23: the rsmA2/A3 double mutant.
Specifically, by comparing the rsmA2/A3 and rsmA3 mutants with the wild-type PstDC3000, a total of 1358 and 1074 DEGs in HMM, and 870 and 1463 DEGs in KB were uncovered, respectively (Supplementary Figures 3A,C,D,F). When comparing the rsmA2/A3 double mutant with the rsmA3 mutant, a total of 277 and 741 DEGs were discovered in HMM and KB, respectively (Supplementary Figure 3B,E), suggesting that more genes were influenced by RsmA2 in KB than in HMM. DEGs were then functionally classified based on COGs. A total of 797, 177, and 601 DEGs in HMM (Supplementary Figure 4A,B,C) and 498, 502, and 949 DEGs in KB (Supplementary Figure 4D,E,F) were functionally categorized into 20 known function categories in the three comparisons (ΔrsmA23 versus PstDC3000, ΔrsmA23 versus ΔrsmA3, and ΔrsmA3 versus PstDC3000), respectively. By comparing the rsmA2/A3 double mutant with the rsmA3 mutant, more DEGs in broad functional categories were found in KB than in HMM (Supplementary Figure 4B,E), further suggesting that RsmA2 might play important roles in KB than in HMM.
To verify the RNA-seq data, seven genes were selected from PstDC3000, including genes encoding catalase (katE), adenylate cyclase (cyaA), transcriptional regulator FleQ (fleQ), phosphate regulon transcriptional regulatory protein (phoB), citrate transporter (citM), sensor histidine kinase (ladS), and pyruvate kinase (pyk). The qRT-PCR results showed that expression of these genes showed a similar trend with those of the RNA-seq data (Supplementary Figure 5).
Expression of rsmX/Y/Z ncsRNAs and rsmAs in PstDC3000 Wild-Type and rsmA Mutant Derivatives
Previous studies revealed that the RsmA family protein positively regulates the transcription of rsmY and rsmZ ncsRNAs in Pseudomonas fluorescens CHA0 (Reimmann et al., 2005). In PstDC3000, RsmA2 and RsmA3 exhibited stronger binding affinities to ncsRNAs (Ge et al., 2019). In this study, rsmX1-5, rsmY, and rsmZ ncsRNAs were down-regulated in the rsmA3 and the rsmA2/A3 double mutants as compared with PstDC3000 (Table 1). Except for rsmY and rsmX4, the FCs of rsmZ, rsmX1, X2, X3, and X5 were much lower in ΔrsmA23 versus PstDC3000 than those in ΔrsmA3 versus PstDC3000 in both HMM and KB media (Table 1). The FCs of rsmY and rsmX4 in both comparisons in both media were similar. These results suggested that rsmY (and possibly rsmX4) were mainly influenced by RsmA3, whereas rsmZ, rsmX1, X2, X3, and X5 were synergistically affected by RsmA2 and RsmA3.
TABLE 1.
Fold changes of the non-coding small RNAs and the rsmA genes in HMM and KB.
| Locus tag | Medium comparison gene/small RNA | HMM ΔrsmA23/ PstDC3000 | HMM ΔrsmA23/Δ rsmA3 | HMM ΔrsmA3/ PstDC3000 | KB ΔrsmA23/PstDC3000 | KB Δ rsmA23/Δ rsmA3 | KB Δ rsmA3/ PstDC3000 |
| PSPTO_5647 | rsmY | −2.09 | / | −1.89 | −2.43 | / | −1.64 |
| PSPTO_5652 | rsmZ | −22.19 | −6.25 | −3.55 | −42.31 | −23.20 | −1.8 |
| PSPTO_5671 | rsmX1 | −9.83 | −3.37 | −2.91 | −91.63 | −8.94 | −10.25 |
| PSPTO_5672 | rsmX2 | −4.26 | −2.7 | −1.57 | −16.63 | / | −9.9 |
| PSPTO_5673 | rsmX3 | −1.43 | −1.48 | / | −1.97 | −1.9 | / |
| PSPTO_5674 | rsmX4 | −14.47 | / | −10.2 | −60.09 | / | −38.81 |
| PSPTO_5675 | rsmX5 | −7.59 | −4.52 | −1.68 | −18.7 | −3.74 | −4.9 |
| PSPTO_1629 | rsmA1 | 2.49 | / | 2.32 | 1.9 | / | 1.57 |
| PSPTO_1844 | rsmA2 | −20 | −15.56 | / | −11.28 | −6.16 | −1.83 |
| PSPTO_3943 | rsmA4 | / | / | / | / | / | / |
| PSPTO_5621 | rsmA5 | 2.28 | / | 1.69 | 1.66 | −2.93 | 4.86 |
Differential expression in ΔrsmA23 versus PstDC3000, ΔrsmA23 versus ΔrsmA3, and ΔrsmA3 versus PstDC3000 with fold change (FC) ≥ 1.5 or FC ≤ −1.5 and p value < 0.05. /: non-differentially expressed genes (DEGs); HMM: hrp-inducing minimum medium; KB: King’s B medium; PstDC3000: Pseudomonas syringae pv. tomato DC3000; ΔrsmA3: the rsmA3 mutant; ΔrsmA23: the rsmA2/A3 double mutant.
On the other hand, expression of the rsmA2 gene was down-regulated in ΔrsmA3 versus PstDC3000, whereas the expression of the rsmA1 and rsmA5 genes was similarly up-regulated in ΔrsmA3 versus PstDC3000 and ΔrsmA23 versus PstDC3000 in both HMM and KB media (Table 1). No change was observed for the rsmA4 gene (Table 1). These results suggested that RsmA3 might positively affect the expression of the rsmA2 gene (Ge et al., 2019) and negatively influence both rsmA1 and rsmA5 gene expression in PstDC3000.
Transcriptomic Analysis Revealed Distinct and Overlapping Gene Regulation by RsmA2 and RsmA3 in PstDC3000
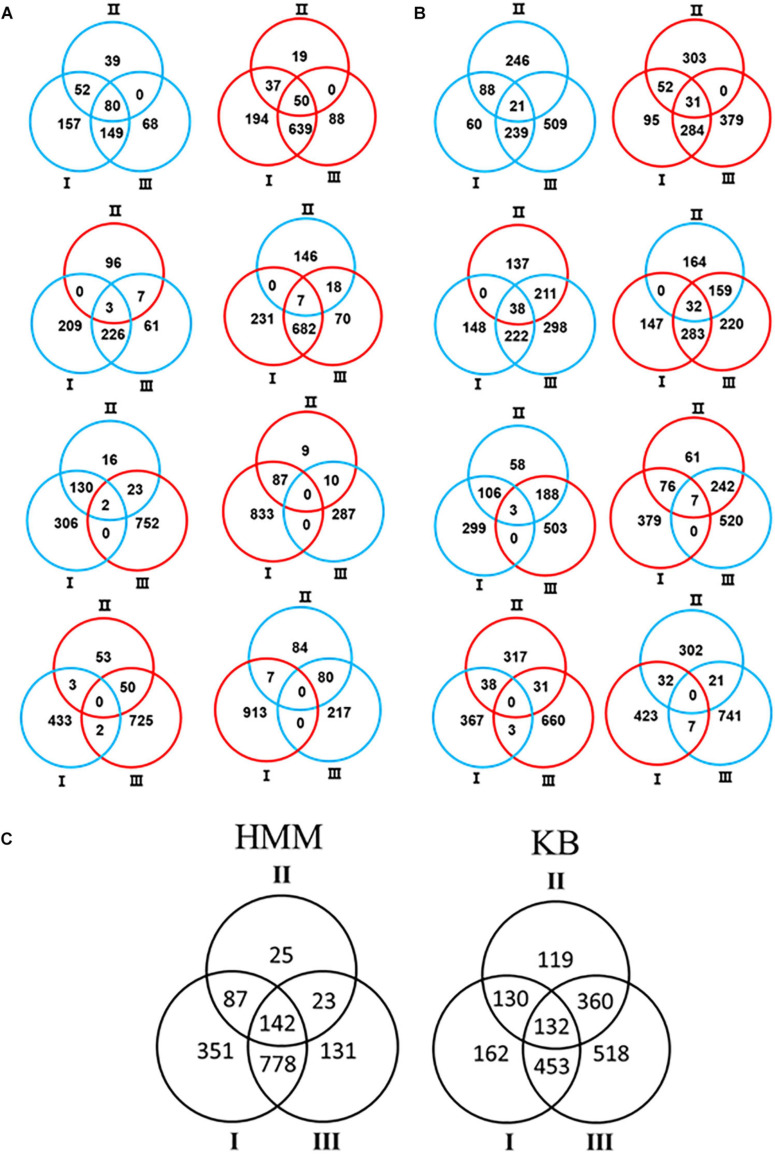
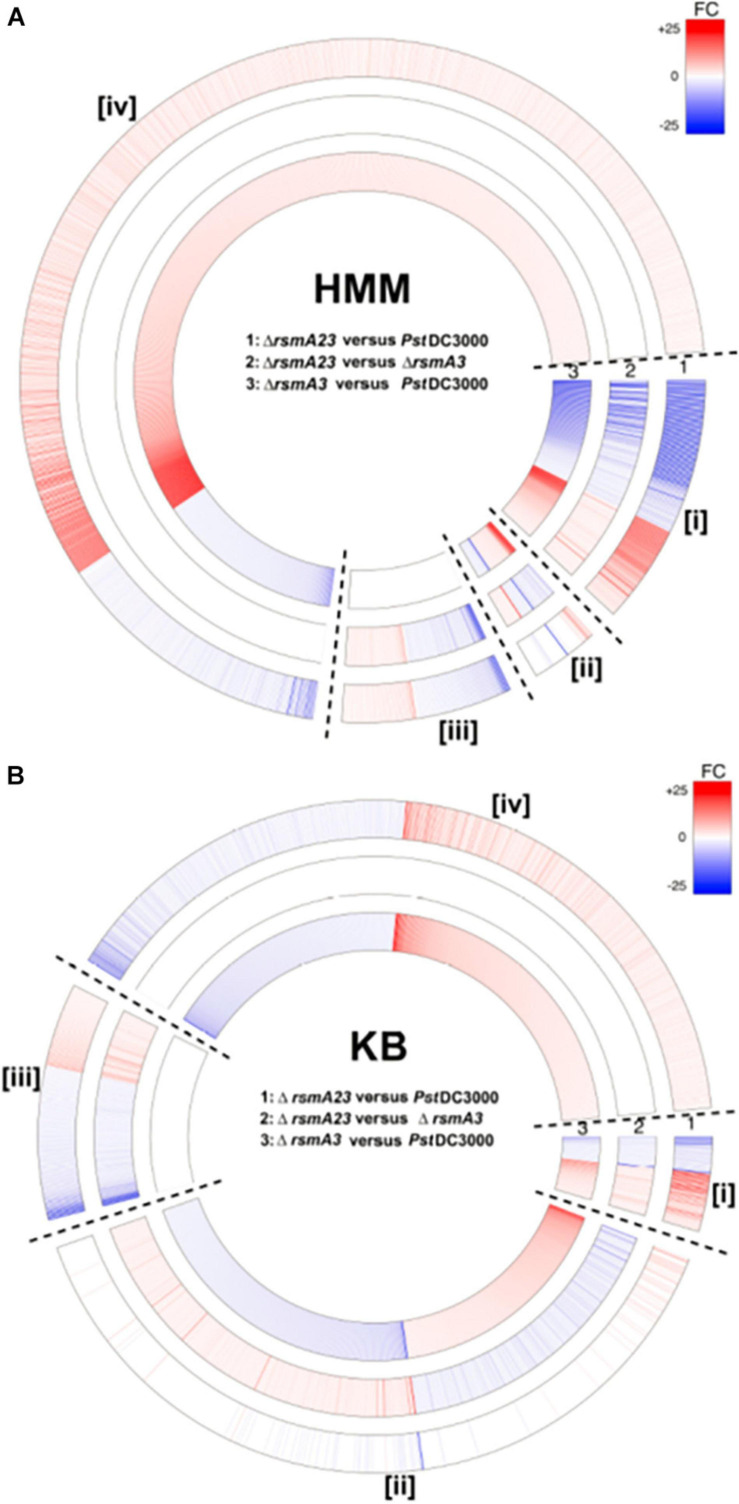
In order to comprehensively understand the differential role of RsmA2 and RsmA3 in regulating gene expression, Venn diagrams were generated to group genes differentially regulated by RsmA2 and RsmA3 in both HMM and KB media (Figure 2). The expression patterns in HMM and KB were further visualized using a circle plot and divided into four major groups (Figure 3). Group [i] includes 130 and 52 genes that were synergistically regulated by RsmA2 and RsmA3 in HMM and KB, respectively (Figure 3). Among them, 80 and 21 were positively affected by RsmA2 and RsmA3 in HMM and KB, respectively, whereas 50 and 31 were negatively influenced by RsmA2 and RsmA3 in HMM and KB, respectively (Figures 2A,B and Table 2). Group [ii] includes 35 and 440 genes that were inversely regulated by RsmA2 and RsmA3 in HMM and KB, respectively (Figure 3). Among them, the expression of 10 and 249 genes was inhibited by RsmA2 but activated by RsmA3 in HMM and KB, respectively (Figures 2A,B and Table 2). In contrast, 25 and 191 genes were activated by RsmA2 but suppressed by RsmA3 in HMM and KB, respectively (Figures 2A,B and Table 2). Group [iii] includes 87 and 130 genes that were mainly influenced by RsmA2 in HMM and KB, respectively (Figures 2C, 3). Among them, 50 and 85 were activated by RsmA2 in HMM and KG, respectively, whereas 37 and 45 genes were inhibited by RsmA2 in HMM and KB, respectively (Table 2). Group [iv] includes 778 and 453 genes that were mainly affected by RsmA3 in HMM and KB, respectively (Figures 2C, 3). Among them, 146 and 201 were activated by RsmA3 in HMM and KB, whereas 632 and 252 genes were suppressed by RsmA3 in HMM and KB, respectively (Table 2).
FIGURE 2.
Venn diagram showed number of differentially expressed genes (DEGs) in each of the three comparisons. (A) HMM. (B) KB. Red and blue circles represent up- and down-regulated genes, respectively. (C) Combination of HMM and KB data. Black circles represent differential expression including both up- and down-regulated genes. Comparison I: ΔrsmA23 versus PstDC3000; II: ΔrsmA23 versus ΔrsmA3; III: ΔrsmA3 versus PstDC3000; HMM: hrp-inducing minimum medium; KB: King’s B medium; PstDC3000: Pseudomonas syringae pv. tomato DC3000; ΔrsmA3: the rsmA3 mutant; ΔrsmA23: the rsmA2/A3 double mutant.
FIGURE 3.
Circular map of differentially expressed genes (DEGs) based on differential regulation by RsmA2 and RsmA3 in PstDC3000. (A) Fold changes (FCs) of ΔrsmA23 versus PstDC3000 (#1), ΔrsmA23 versus ΔrsmA3 (#2), and ΔrsmA3 versus PstDC3000 (#3) in HMM. (B) FCs of ΔrsmA23 versus PstDC3000 (#1), ΔrsmA23 versus ΔrsmA3 (#2), and ΔrsmA3 versus PstDC3000 (#3) in KB. Group [i] represents genes synergistically regulated by RsmA2 and RsmA3; group [ii] represents genes inversely regulated by RsmA2 and RsmA3; group [iii] represents genes regulated mainly by RsmA2; and group [iv] represents genes regulated mainly by RsmA3 (see Table 2). DEGs were defined as genes with a | Fold Change (FC)| value ≥ 1.5 and a p value < 0.05. Up- and down-regulated DEGs were indicated by red and blue, respectively. The higher the | FC|, the deeper the color. HMM: hrp-inducing minimum medium; KB: King’s B medium; PstDC3000: Pseudomonas syringae pv. tomato DC3000; ΔrsmA3: the rsmA3 mutant; ΔrsmA23: the rsmA2/A3 double mutant.
TABLE 2.
Numbers of differentially expressed genes (DEGs) based on regulation by RsmA2 and RsmA3 in PstDC3000.
| HMM | KB | |||||
| Groups | Down | Up | Total | Down | Up | Total |
| [i] Genes synergistically regulated by RsmA2 and RsmA3 | 80 | 50 | 130 | 21 | 31 | 52 |
| [ii] Genes inversely regulated by RsmA2 and RsmA3 | 10 | 25 | 35 | 249 | 191 | 440 |
| [iii] Genes regulated mainly by RsmA2 | 50 | 37 | 87 | 85 | 45 | 130 |
| [iv] Genes regulated mainly by RsmA3 | 146 | 632 | 778 | 201 | 252 | 453 |
The number of genes was generated from Figure 3. Down and up represent expression in comparisons, i.e., ΔrsmA3 versus PstDC3000, ΔrsmA23 versus PstDC3000, or ΔrsmA23 versus ΔrsmA3. [i] DEGs up-regulated (or down-regulated) simultaneously in all three comparisons. [ii] DEGs inversely expressed in ΔrsmA23 versus ΔrsmA3 and ΔrsmA3 versus PstDC3000; down or up represents expression in ΔrsmA3 versus PstDC3000. [iii] DEGs in ΔrsmA23 versus PstDC3000 and ΔrsmA23 versus ΔrsmA3, but not in ΔrsmA3 versus PstDC3000. [iv] DEGs in ΔrsmA23 versus PstDC3000 and ΔrsmA3 versus PstDC3000, but not in ΔrsmA23 versus ΔrsmA3. HMM: hrp-inducing minimum medium; KB: King’s B medium; PstDC3000: Pseudomonas syringae pv. tomato DC3000; ΔrsmA3: the rsmA3 mutant; ΔrsmA23: the rsmA2/A3 double mutant.
Overlapping and Distinct Gene Regulation by RsmA2 and RsmA3 in HMM
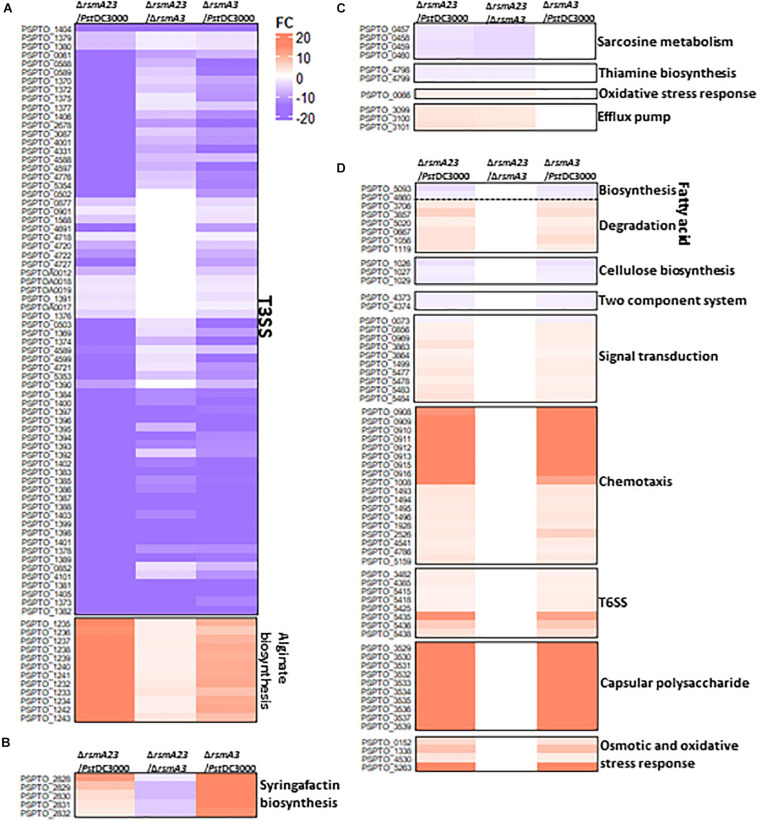
In HMM, about 130 genes were synergistically regulated by RsmA2 and RsmA3 (Figure 3A[i] and Supplementary Table 2). Specifically, both RsmA2 and RsmA3 activated 71 T3SS-related genes and inhibited 12 genes involved in alginate biosynthesis in a synergistic way (Figure 4A and Supplementary Table 4). On the other hand, about 35 genes were inversely regulated by RsmA2 and RsmA3 in HMM (Figure 3A[ii] and Supplementary Table 2). Among them, genes related with syringafactin biosynthesis (syrR, syfABCD) were up-regulated in ΔrsmA3 versus PstDC3000 but down-regulated in ΔrsmA23 versus ΔrsmA3, suggesting that RsmA3 negatively and RsmA2 positively influenced syringafactin gene expression (Figure 4B and Supplementary Table 5). About 87 genes were regulated mainly by RsmA2 in HMM (Figure 3A[iii] and Supplementary Table 2). Among them, 50 genes were activated mainly by RsmA2, including the sox gene cluster involved in sarcosine metabolism and the thiD and thiE genes involved in thiamine biosynthesis (Figure 4C and Supplementary Table 6); whereas 37 genes were suppressed mainly by RsmA2, including genes involved in efflux pump (mexE, saxF, and oprN) (Figure 4C). Furthermore, about 778 genes were regulated mainly by RsmA3 in HMM, including 146 activated genes and 632 suppressed genes (Figure 3A[iv] and Supplementary Table 2). Genes involved in fatty acid metabolism, cellulose synthases (wssABD), and two-component regulatory system (TCRS, ColRS) were activated mainly by RsmA3, whereas genes involved in signal transduction (spoT, pspto_0856, and dksA), QS (psyR and psyI), c-di-GMP (wspR), phosphate metabolism, type VI secretion system (T6SS), and stress responses were inhibited mainly by RsmA3 (Figure 4D and Supplementary Table 7).
FIGURE 4.
Heat maps of selected differentially expressed genes (DEGs) in HMM. (A) T3SS and alginate biosynthesis genes synergistically regulated by RsmA2 and RsmA3. (B) Syringafactin biosynthesis genes inversely regulated by RsmA2 and RsmA3. (C) Genes regulated mainly by RsmA2. (D) Genes regulated mainly by RsmA3. DEGs were defined as genes with a | Fold Change (FC)| value ≥ 1.5 and a p value < 0.05. Up- and down-regulated DEGs were indicated by red and blue, respectively. The higher the | FC|, the deeper the color. HMM: hrp-inducing minimum medium; PstDC3000: Pseudomonas syringae pv. tomato DC3000; ΔrsmA3: the rsmA3 mutant; ΔrsmA23: the rsmA2/A3 double mutant; T3SS, type III secretion system; T6SS, type VI secretion system.
Overlapping and Distinct Gene Regulation by RsmA2 and RsmA3 in KB
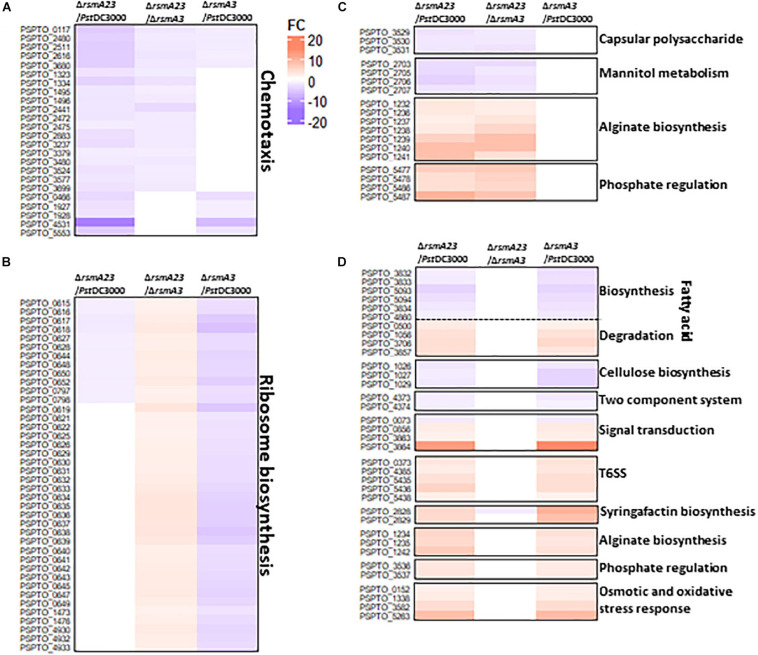
Similarly, about 52 genes were synergistically regulated by RsmA2 and RsmA3 in KB (Figure 3B[i] and Supplementary Table 3). Both RsmA3 and RsmA2 synergistically activated 24 chemotaxis-related genes and suppressed the transcription of alginate biosynthesis genes, including algA, algL, algX, algG, algE, algK, alg44, algJ, algI, alg8, and algD genes (Figures 5A,C,D and Supplementary Tables 8,10, 11), whereas 440 genes were inversely regulated by RsmA2 and RsmA3 in KB (Figure 3B[ii] and Supplementary Table 3). As an example, 40 genes encoding ribosomal proteins were down-regulated in ΔrsmA3 versus PstDC3000 but up-regulated in ΔrsmA23 versus ΔrsmA3, suggesting that RsmA3 activates and RsmA2 suppresses ribosomal gene expression (Figure 5B and Supplementary Table 9). In addition, about 130 genes were regulated mainly by RsmA2 in KB, including 85 activated and 45 inhibited genes (Figure 3B[iii] and Supplementary Table 3). Among them, the uxuB and mannitol ABC transporter genes involved in mannitol metabolism were activated, whereas genes related with phosphate regulation were suppressed (Figure 5C and Supplementary Table 10). On the other hand, 201 and 252 genes were activated and inhibited mainly by RsmA3 in KB, respectively (Figure 3B[iv] and Supplementary Table 3). Consistent with HMM, genes encoding fatty acid metabolism-related proteins, the wssABD and colRS genes were activated mainly by RsmA3, whereas genes involved in signal transductions [(p)ppGpp and QS], T6SS, and stress responses were suppressed mainly by RsmA3 (Figure 5D and Supplementary Table 11). However, the pslABD genes were activated by RsmA2 and the pslIJ genes were inhibited by RsmA3 in KB, indicating that the capsular polysaccharide (CPS) was regulated by RsmA2 and RsmA3 differently (Figures 5C,D and Supplementary Tables 10, 11).
FIGURE 5.
Heat maps of selected differentially expressed genes (DEGs) in KB. (A) Chemotaxis genes synergistically regulated by RsmA2 and RsmA3. (B) Genes encoding ribosome proteins inversely regulated by RsmA2 and RsmA3. (C) Genes regulated mainly by RsmA2. (D) Genes regulated mainly by RsmA3. DEGs were defined as genes with a | Fold Change (FC)| value ≥ 1.5 and a p value < 0.05. Up- and down-regulated DEGs were indicated by red and blue, respectively. The higher the | FC|, the deeper the color. KB: King’s B medium; PstDC3000: Pseudomonas syringae pv. tomato DC3000; ΔrsmA3: the rsmA3 mutant; ΔrsmA23: the rsmA2/A3 double mutant; T6SS, type VI secretion system.
Syringafactin Production and Oxidative Stress Resistance Were Negatively Regulated by RsmA3 in PstDC3000
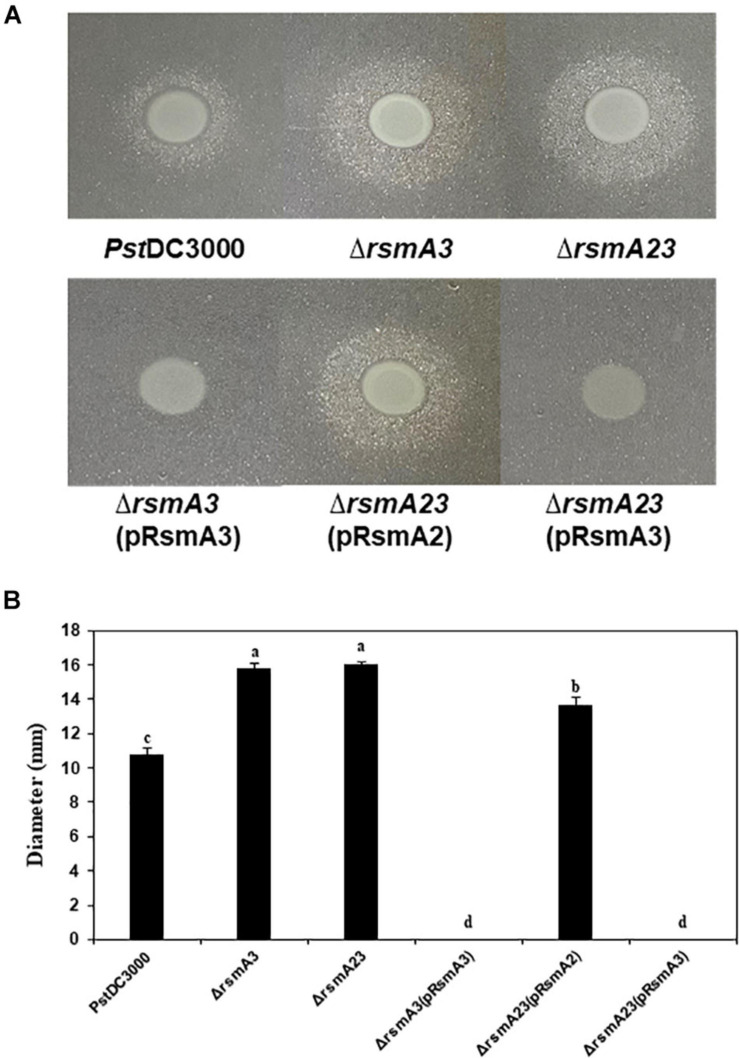
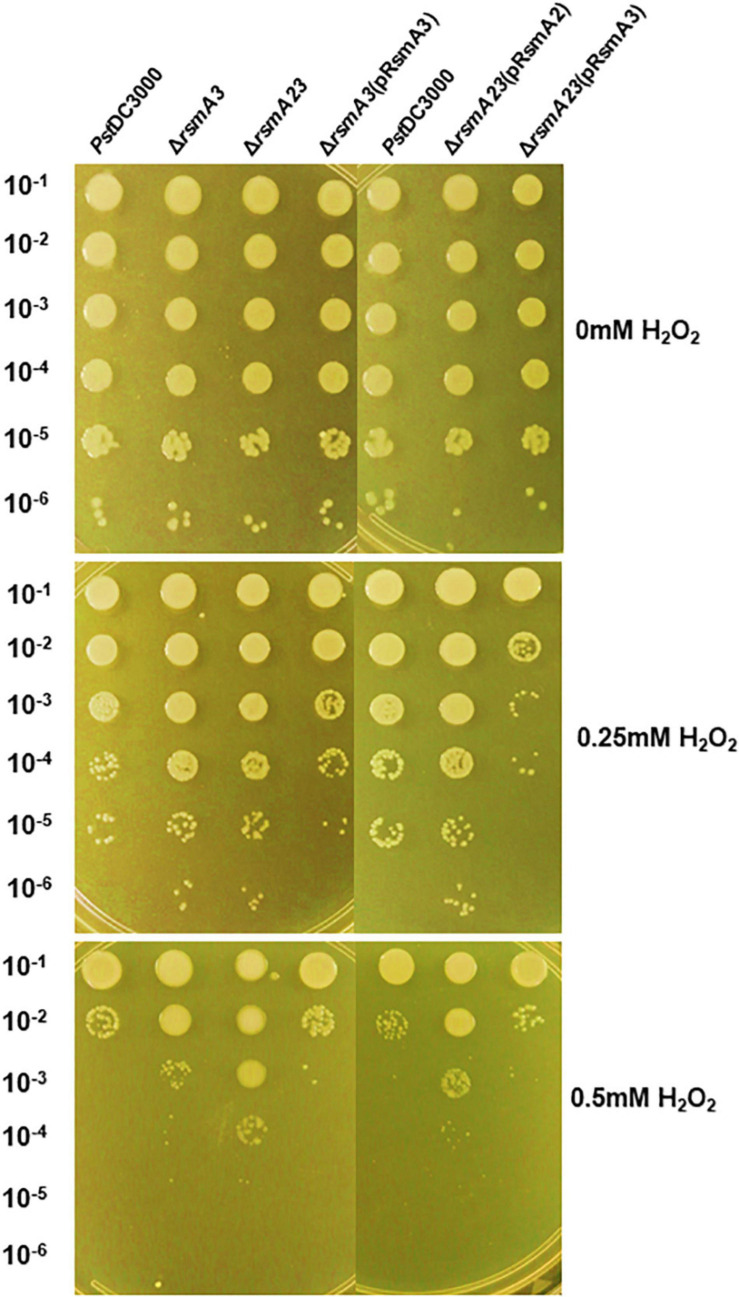
To confirm the effect of RsmA3 on syringafactin biosynthesis in KB, atomized oil assay was used to measure syringafactin production in PstDC300 and its derived rsmA mutant strains. Both the rsmA3 and the rsmA2/A3 double mutants showed significantly increased production of syringafactin as compared with the wild-type PstDC3000 (Figure 6). However, expression of the rsmA3 gene in both the rsmA3 mutant and the rsmA2/A3 double mutant led to almost no syringafactin production (Figure 6). These results indicated that RsmA3 strongly affected syringafactin production in a negative way in PstDC3000 and RsmA2 might play a minor role. In addition, to confirm the role of RsmA3 in response to oxidative stress, spot dilution assay results showed that both the rsmA3 and the rsmA2/A3 double mutants exhibited increased oxidative resistance as compared with PstDC3000 (Figure 7). Complementation of the mutants with the rsmA3 gene partially restored oxidative resistance of the rsmA3 mutant to the wild-type level (Figure 7). In addition, complementation of the rsmA23 mutants, i.e., rsmA23 (pRsmA2) and rsmA23 (pRsmA3), restored to the rsmA3 mutant and wild-type level, respectively (Figure 7). These results suggest that RsmA3 negatively regulates antioxidant stress in PstDC3000 and RsmA2 might play a minor role.
FIGURE 6.
Syringafactin production in PstDC3000 and its mutants and complementation stains. (A) Syringafactin halos around PstDC3000 and its derived stains. (B) Diameter of syringafactin halos around PstDC3000 and its derived strains. Bacteria were grown on KB plates (1.5% agar) for 24 h at 20°C before a mist of mineral oil was sprayed over. Halo diameters were measured to represent syringafactin production. Different letters in panel (B) indicate significant difference from one-way ANOVA followed by Fisher’s LSD test (p < 0.05). Error bars represented standard deviations. PstDC3000: Pseudomonas syringae pv. tomato DC3000; ΔrsmA2: the rsmA2 mutant; ΔrsmA3: the rsmA3 mutant; ΔrsmA23: the rsmA2/A3 double mutant; pRsmA2: pUCP18 containing the rsmA2 gene with native promoter; pRsmA3: pUCP18 containing the rsmA3 gene with native promoter.
FIGURE 7.
Sensitivity to oxidative stress of PstDC3000 and its derived strains. Serial 10-fold dilutions were made from OD600 (optical density at 600 nm) = 0.1 in KB. Five microliters of each dilution were added to KB agar plates amended with 0, 0.25, and 0.5 mM H2O2. Photographs were taken 2 days post-incubation. PstDC3000: Pseudomonas syringae pv. tomato DC3000; ΔrsmA3: the rsmA3 mutant; ΔrsmA23: the rsmA2/A3 double mutant; pRsmA2: pUCP18 containing the rsmA2 gene with native promoter; pRsmA3: pUCP18 containing the rsmA3 gene with native promoter.
Discussion
The CsrA/RsmA RNA-binding protein is a global dual post-transcriptional regulator (Romeo et al., 2013; Vakulskas et al., 2015). Bacteria usually possess one or two homologous CsrAs/RsmAs (Pessi et al., 2001; Chatterjee et al., 2003; Ancona et al., 2016). However, five or even seven CsrAs/RsmAs are present in the P. syringae genome (Ge et al., 2019; Ramírez-Zapata et al., 2020; Sobrero and Valverde, 2020), and these CsrA/RsmA homologs appear to be functional. It has been further reported that CsrAs/RsmAs in P. syringae played redundant or unique roles, especially RsmA2 (RsmA) and RsmA3 (RsmE) in PstDC3000 in regulating T3SS, alginate production, motility, and protease activities (Ferreiro et al., 2018; Ge et al., 2019; Ramírez-Zapata et al., 2020). In this study, comparative transcriptomic analysis established that RsmA proteins exhibited distinct and overlapping roles in modulating virulence and survival in P. syringae under different nutritional conditions and shed light on the potential interplay of the RsmA proteins. We further confirmed as how RsmA2 and RsmA3 regulated syringafactin production and oxidative stress resistance in PstDC3000.
The interplay among the RsmA homologs has not been intensively investigated in plant-associated pseudomonads. In P. fluorescens, expression of RsmE was inhibited by both RsmA and RsmE, the later negatively regulated itself (Reimmann et al., 2005). In Pseudomonas putida, RsmA and RsmE negatively regulated their own expression (Huertas-Rosales et al., 2016). Furthermore, RsmA binds to rsmA and rsmF mRNA in vitro to repress RsmA and RsmF translation in P. aeruginosa (Marden et al., 2013). In this study, we found that RsmA3 positively affects the rsmA2 gene expression and negatively influences both rsmA1 and rsmA5 gene expression. This is consistent with previous report that RsmA3 (RsmE) in PstDC3000 promoted the expression of RsmA2 (RsmA) (Ge et al., 2019). In addition, we previously demonstrated that the RsmA2, RsmA3, and RsmA4 protein levels were much lower in rsmA2 and rsmA3 double mutants than in the rsmA3 single mutant, further suggesting that RsmA2 and RsmA3 might synergistically influence the expression of RsmA2, RsmA3, and RsmA4 proteins and RsmA2 may also reciprocally influence RsmA3 expression at the post-transcriptional level (Ge et al., 2019). These results suggest that cross talk between RsmAs in PstDC3000 is very complicated and RsmA3 might be on the top of the regulatory system by controlling other RsmAs at transcriptional, post-transcriptional, and translational levels. On the other hand, GacS and GacA positively control the expression of ncsRNAs (Cui et al., 2001; Chatterjee et al., 2003; Ramírez-Zapata et al., 2020), which sequester and antagonize CsrA/RsmA activities. Earlier studies identified seven ncsRNAs, i.e., rsmX1-5, rsmZ, and rsmZ, in the PstDC3000 genome (Moll et al., 2010), and these ncsRNAs strongly bind to RsmA2 and RsmA3, but less to RsmA1 and RsmA4 (Ge et al., 2019). In this study, we showed that both RsmA2 and RsmA3 positively affect the transcription of rsmX1, X2, X3, X5, and rsmZ ncsRNAs, whereas the expression of rsmY (and possibly rsmX4) is positively influenced mainly by RsmA3. Similar results have also been reported in E. amylovora (Ancona et al., 2016). These results suggest that RsmA3 together with RsmA2 might influence the gacS/gacA mRNAs, thus positively regulating rsmX/Y/Z ncsRNAs in PstDC3000 as a negative feedback loop, or directly affect the stability of ncsRNAs (Ge et al., 2019).
It has been previous reported that single rsmA1/A2/A3/A4 mutation in PstDC3000 did not affect virulence, while the rsmA2/A3 double mutant exhibited significantly reduced virulence and bacterial growth in planta (Ge et al., 2019). It is thus hypothesized that genes regulated by both RsmA2 and RsmA3 might be important for bacterial growth and virulence. The T3SS is a key virulence factor in PstDC3000 by suppressing host defense through injecting effectors into host cells, and the T3SS genes are directly regulated by the RpoN-HrpL sigma factor cascade (Zhao et al., 2003; Schechter et al., 2004). In E. amylovora, CsrA promoted the expression of the T3SS gene, which is also directly regulated by the RpoN-HrpL sigma factor cascade (Ancona et al., 2014, 2016; Lee et al., 2019). In this study, RsmA2 and RsmA3 synergistically activated 71 T3SS-related genes in HMM, including hrpL and hrpR/S, suggesting that RsmA2 and RsmA3 might target similar upstream regulatory genes to influence T3SS genes. In X. citri, RsmA protects the hrpG transcript, encoding the master regulator of T3SS, from RNase E cleavage and allows T3SS gene expression (Andrade et al., 2014). However, both HrpL and HrpS were not direct targets of CsrA in E. amylovora, for which the relA mRNA is a direct target (Lee et al., 2019). It is reasonable to speculate that RsmA2/RsmA3 might also directly target upstream regulatory genes such as relA mRNA to regulate T3SS in PstDC3000.
Exopolysaccharides in P. syringae play a key role in bacterial survival and virulence under stress conditions by avoiding recognition of host plant, resisting to desiccation, and enhancing epiphytic fitness (Lindow, 1991; Kasapis et al., 1994). Alginate is a major EPS in P. syringae and P. aeruginosa (Penaloza-Vazquez et al., 1997, 2004; Ramsey and Wozniak, 2005). It has previously been reported that the csrA3, but not csrA2, mutant significantly increased the production of alginate in both LB and minimal medium and reduced the expression of the algD gene, the first gene in the alginate biosynthesis operon (Ferreiro et al., 2018). In contrast, we found that both RsmA2 and RsmA3 negatively influenced the expression of the alginate biosynthesis genes as reported previously (Ge et al., 2019). This discrepancy might be due to the fact that RsmA3 positively affects rsmA2 expression, and the role of RsmA2 in alginate biosynthesis might only be evident when RsmA3 is absent. On the other hand, products of the psl gene cluster involved in CPS biosynthesis is essential for biofilm formation in P. aeruginosa (Overhage et al., 2005; Campisano et al., 2006), whereas cellulose impacts biofilm formation at the air–liquid interface (Pérez-Mendoza et al., 2014; Prada-Ramírez et al., 2016). In PstDC3000, the psl gene cluster was suppressed mainly by RsmA3 in HMM. However, the psl genes in KB were activated and suppressed by RsmA2 and RsmA3, respectively. In contrast, cellulose biosynthesis genes were activated mainly by RsmA3 in both HMM and KB.
Many bacteria use flagella-driven motility to respond to changes in their chemical environment, a process known as chemotaxis (Berg and Brown, 1972; Wadhams and Armitage, 2004; Clarke et al., 2016). In P. aeruginosa, chemotaxis pathway-related genes play an important role in flagellar motility, T4 pili formation, and biofilm formation (Hickman et al., 2005). In Pseudomonas savastanoi and PstDC3000, chemotaxis plays a role in virulence (Matas et al., 2012; Clarke et al., 2016) and the rsmA2/rsmA3 mutant of PstDC3000 showed significantly reduced motility (Ge et al., 2019). It is well established that CsrA binding of flhD inhibits RNase E-mediated cleavage and enhances its translation rate in E. coli (Wei et al., 2001; Yakhnin et al., 2013). In E. amylovora, CsrA also binds to multiple sites on the flhD transcript, resulting in enhanced translation (Lee et al., 2019). Here we showed that expression of the chemotaxis-related genes was positively influenced by both RsmA2 and RsmA3. It is plausible that reduced motility in the rsmA2/rsmA3 mutant was partly due to reduced expression of chemotaxis genes and FleQ, the regulator for flagellar biosynthesis in P. syringae (Nogales et al., 2015), might be a direct target of RsmA2/3, which requires further investigation.
On the other hand, bacterial pathogens produce biosurfactants such as syringafactin to enhance swarming motility and acquire nutrients on leaf surfaces (Burch et al., 2014; Nogales et al., 2015). Syringafactin produced by P. syringae promotes bacterial proliferation by increasing the permeability of cuticle and the hygroscopicity of water to improve intake of internal nutrients (Burch et al., 2014). In this study, syringafactin biosynthesis genes were inversely influenced by RsmA3 and RsmA2, suggesting that syringafactin production is tightly regulated to fine-tune bacterial motility on leaf surface. Furthermore, the transcription factor OxyR controls the catalase-related genes (katB, katE, and katG) to cope with plant reactive oxidative stress (ROS) response in PstDC3000 (Ishiga and Ichinose, 2016). Oxidative stress response genes including the oxyR gene and the katB, katE, and katG genes in HMM were inhibited by RsmA2 and RsmA3, respectively, whereas osmotic stress response gene osmC was also suppressed by RsmA3. These results suggest that RsmA proteins in PstDC3000 play important roles in modulating stress responses for survival.
As a global post-transcriptional regulator in bacteria, CsrA/RsmA controls more than 10% of total genes in Salmonella typhimurium and P. aeruginosa (Lawhon et al., 2003; Burrowes et al., 2006). It was also revealed that CsrA could directly interact with about 10% of total gene transcripts in S. typhimurium, Legionella pneumophila, and Campylobacter jejuni (Dugar et al., 2016; Holmqvist et al., 2016; Sahr et al., 2017). In E. coli, about 25 and 12.5% of total gene transcripts were differentially expressed in the csrA mutant and directly bound by CsrA in vivo, respectively (Potts et al., 2017). In E. amylovora, CsrA affects about 20% of the genes in the genome (Lee et al., 2019). In this study, RsmA3 modulated about 20 and 27% of genes in the PstDC3000 genome in HMM and KB, respectively. Considering both RmsA3 and RsmA2, the percentage of genes affected increased to 29 and 35% in HMM and KB, respectively. When all three comparisons in both conditions were considered, about half (49%) of genes in the genome was differentially expressed. It is reasonable to speculate that RsmAs in PstDC3000 might directly bind to their targets or indirectly affect gene expression through global regulatory systems. Expression of global regulatory systems affected in this study includes secondary messengers (p)ppGpp, c-di-GMP, QS (Las and Rhl QS), and TCSs (GacS/GacA; ColR/ColS) (Stock et al., 2000; Miller and Bassler, 2001; Pessi et al., 2001; Kalia et al., 2013; Liu et al., 2020), suggesting that global regulatory systems might be targets for RsmA proteins.
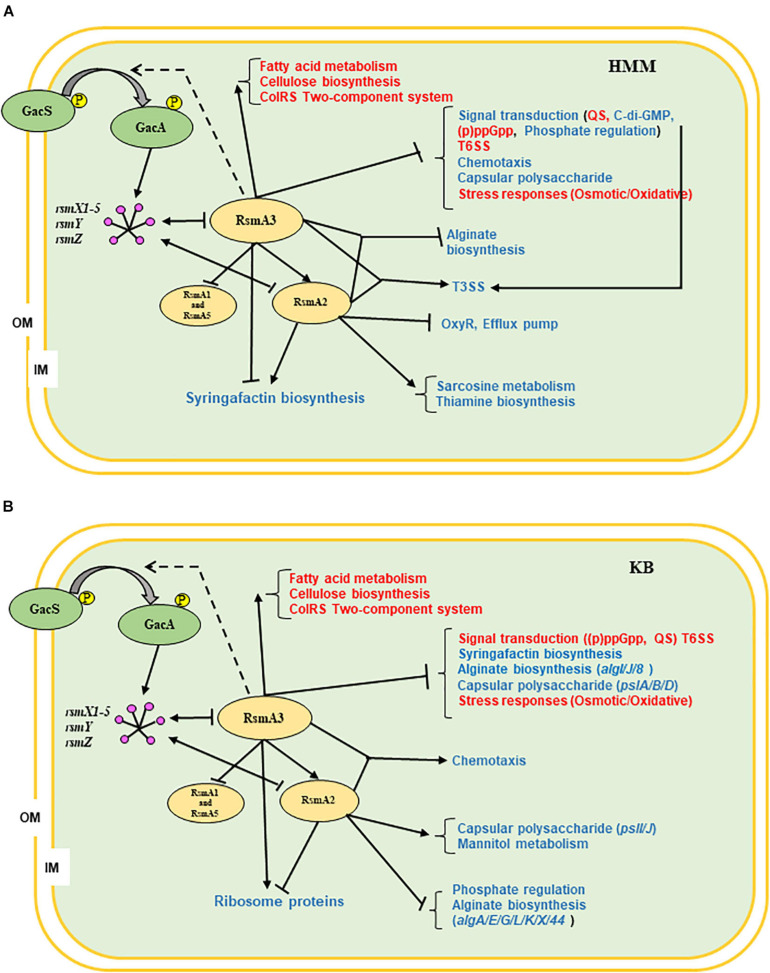
In summary, we propose the following working models for RsmA proteins in modulating gene expression in PstDC3000 based on current data (Figure 8). RsmA3 activates the rsmA2 gene expression and suppresses both rsmA1 and rsmA5 gene expression at transcriptional level. RsmA2 or RsmA3 in turn affected the expression levels of the rsmY, rsmZ, and rsmX1-5 ncsRNAs. RsmA2 and RsmA3 synergistically control the expression of genes involved in T3SS and alginate biosynthesis in HMM, and chemotaxis in KB, whereas RsmA2 and RsmA3 inversely modulate transcripts of genes involved in syringafactin production in HMM, and ribosomal protein biosynthesis in KB. In addition, both RsmA2 and RsmA3 play a major role in influencing specific or conserved pathway genes in different nutritional environments as summarized in Figure 8 (red/blue texts), indicating their potential direct targets. In the future, researches should focus more on determining the impacts of the other RsmA homologs or overall impacts of all RsmAs, the precise interplay among all five RsmA homologs, and specifically the molecular targets of RsmAs in PstDC3000.
FIGURE 8.
Proposed working models illustrating the global effects of RsmA2 and RsmA3 in PstDC3000 as well as potential interplay among RsmA proteins. (A) HMM. (B) KB. GacS/GacA: two-component regulatory systems; rsmX1-X5, rsmY, rsmZ: small non-coding regulatory RNAs (sncRNAs); RsmA1, RsmA2, RsmA3, and RsmA5: RNA-binding proteins; OM: outer membrane; IM: inner membrane; ↓: positive effect; ⊥: negative effect; dashed line: unknown. Red and blue fonts represent similarity and difference between KB and HMM, respectively. HMM: hrp-inducing minimum medium; KB: King’s B medium; PstDC3000: Pseudomonas syringae pv. tomato DC3000; (p)ppGpp: guanosine tetra/pentaphosphate; QS: quorum sensing; T3SS: type III secretion system; T6SS: type VI secretion system.
Data Availability Statement
RNA-seq data files have been submitted to Gene Expression Omnibus (GEO) at the National Center for Biotechnology Information (NCBI) with an accession number GSE162091.
Author Contributions
YZ and BH designed the research. JL, MY, YG, and YT performed the research and analyzed the data. JL, MY, and YZ wrote the manuscript. All authors have read and approved the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
JL and YG would like to thank the financial support from the China Scholarship Council.
Funding. This project was supported by the Agriculture and Food Research Initiative Competitive Grants Program Grant No. 2016-67013-24812 from the USDA National Institute of Food and Agriculture, USDA-SCRI and USDA-Hatch Project ILLU-802-913 (YZ). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.637595/full#supplementary-material
Multidimensional clustering plot. Dimension 1 and Dimension 2 explained data variability of 60% (media) and 21% (strains), respectively, for strains of PstDC3000, ΔrsmA3, and ΔrsmA23 cultured in HMM and KB media. HMM: hrp-inducing minimum medium; KB: King’s B medium; PstDC3000: Pseudomonas syringae pv. tomato DC3000; ΔrsmA3: the rsmA3 mutant; ΔrsmA23: the rsmA2/A3 double mutant.
Heatmap. (A) PstDC3000, ΔrsmA3, and ΔrsmA23 in both HMM and KB, with a total of 2661 differentially expressed genes (DEGs). (B) PstDC3000, ΔrsmA3, and ΔrsmA23 in HMM with a total of 1560 DEGs. (C) PstDC3000, rsmA3, and rsmA23 in KB with a total of 1879 DEGs. Up- and down-regulated genes were indicated by red and blue colors, respectively. HMM: hrp-inducing minimum medium; KB: King’s B medium; PstDC3000: Pseudomonas syringae pv. tomato DC3000; ΔrsmA3: the rsmA3 mutant; ΔrsmA23: the rsmA2/A3 double mutant.
MA plots (a.k.a., mean-difference plots; M: log2FC; A: average log2CPM; CPM: counts per million reads). (A) ΔrsmA23 versus PstDC3000 in HMM. (B) ΔrsmA23 versus ΔrsmA3 in HMM. (C) ΔrsmA3 versus PstDC3000 in HMM. (D) ΔrsmA23 versus PstDC3000 in KB. (E) ΔrsmA23 versus ΔrsmA3 in KB. (F) ΔrsmA3 versus PstDC3000 in KB. M: log2FC, A: Average Log2CPM, counts per million reads. Dots between two purple lines represent | log2FC| value ≤ 0.58496250072 (i.e., | FC| value ≤ 1.5), and outside dots represent | log2FC| value ≥ 0.58496250072 (i.e., | FC| value ≥ 1.5). Up- and down-regulated genes were indicated by red and blue dot with p value < 0.05, black dot represents no significant difference with p value > 0.05. HMM: hrp-inducing minimum medium; KB: King’s B medium; PstDC3000: Pseudomonas syringae pv. tomato DC3000; ΔrsmA3: the rsmA3 mutant; ΔrsmA23: the rsmA2/A3 double mutant.
Functional classification of differentially expressed genes (DEGs) based on the clusters of orthologous groups (COGs). (A) ΔrsmA23 versus PstDC3000 in HMM. (B) ΔrsmA23 versus ΔrsmA3 in HMM. (C) ΔrsmA3 versus PstDC3000 in HMM. (D) ΔrsmA23 versus PstDC3000 in KB. (E) ΔrsmA23 versus ΔrsmA3 in KB. (F) ΔrsmA3 versus PstDC3000 in KB. DEGs were defined as genes with a | Fold Change (FC)| value ≥ 1.5 and a p value < 0.05. Up- and down-regulated DEGs were indicated by red and blue bars, respectively. HMM: hrp-inducing minimum medium; KB: King’s B medium; PstDC3000: Pseudomonas syringae pv. tomato DC3000; ΔrsmA3: the rsmA3 mutant; ΔrsmA23: the rsmA2/A3 double mutant.
Verification of selected differentially expressed genes (DEGs) using quantitative real-time PCR. (A) Expression of the katE, cyaA, fleQ, phoB, citM, ladS, and pyk genes in the rsmA3 mutant and the rsmA2/A3 double mutant as compared with PstDC3000 grown in HMM. (B) Expression of the katE, cyaA, fleQ, phoB, citM, ladS, and pyk genes in the rsmA3 mutant and the rsmA2/A3 double mutant as compared with PstDC3000 in KB. The values on top of bars represent fold changes of the gene in ΔrsmA23 or ΔrsmA3 versus PstDC3000, while the absence of a value represents no significant difference in the RNA-seq data. HMM: hrp-inducing minimum medium; KB: King’s B medium; PstDC3000: Pseudomonas syringae pv. tomato DC3000; ΔrsmA3: the rsmA3 mutant; ΔrsmA23: the rsmA2/A3 double mutant.
Primers for qRT-PCR used in this study.
List of differentially expressed genes (DEGs) in PstDC3000, the rsmA3 mutant, and the rsmA2/A3 double mutant in HMM.
List of differentially expressed genes (DEGs) in PstDC3000, the rsmA3 mutant, and the rsmA2/A3 double mutant in KB.
Fold changes of the type III secretion system (T3SS) and alginate biosynthesis genes in HMM.
Fold changes of the syringafactin biosynthesis genes in HMM.
Selected genes regulated mainly by RsmA2 in HMM.
Selected genes regulated mainly by RsmA3 in HMM.
Fold changes of the chemotaxis genes in KB.
Fold changes of the ribosome genes in KB.
Selected genes regulated mainly by RsmA2 in KB.
Selected genes regulated mainly by RsmA3 in KB.
References
- Alarcón-Chaidez F. J., Keith L., Zhao Y., Bender C. L. (2003). RpoN (54) is required for plasmid-encoded coronatine biosynthesis in Pseudomonas syringae. Plasmid 49 106–117. 10.1016/s0147-619x(02)00155-5 [DOI] [PubMed] [Google Scholar]
- Ancona V., Lee J. H., Zhao Y. F. (2016). The RNA-binding protein CsrA plays a central role in positively regulating virulence traits in Erwinia amylovora. Sci. Rep. 6:37195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ancona V., Li W. T., Zhao Y. F. (2014). Alternative sigma factor RpoN and its modulator protein YhbH are indispensable for Erwinia amylovora virulence. Mol. Plant Pathol. 15 58–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade M. O., Farah C. S., Wang N. (2014). The post-transcriptional regulator rsmA/csrA activates T3SS by stabilizing the 5’ UTR of hrpG, the master regulator of hrp/hrc genes, in Xanthomonas. PLoS Pathog. 10:e1003945. 10.1371/journal.ppat.1003945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg H. C., Brown D. A. (1972). Chemotaxis in Escherichia coli analyzed by three dimensional tracking. Nature 239 500–504. 10.1038/239500a0 [DOI] [PubMed] [Google Scholar]
- Boch J., Joardar V., Gao L., Robertson T. L., Lim M., Kunkel B. N. (2002). Identification of Pseudomonas syringae pv. tomato genes induced during infection of Arabidopsis thaliana. Mol. Microbiol. 44 73–88. 10.1046/j.1365-2958.2002.02877.x [DOI] [PubMed] [Google Scholar]
- Brooks D. M., Hernández-Guzmán G., Kloek A. P., Alarcón-Chaidez F., Rangaswamy A., Peñaloza-Vázquez A., et al. (2004). Identification and characterization of a well-defined series of coronatine biosynthetic mutan ts of Pseudomonas syringae pv. tomato strain DC3000. Mol. Plant Microbe Interact. 17 162–174. 10.1094/mpmi.2004.17.2.162 [DOI] [PubMed] [Google Scholar]
- Buell C. R., Joardar V., Lindeberg M., Selengut J., Paulsen I. T., Giglio M., et al. (2003). The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. U.S.A. 100 10181–10186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch A. Y., Shimada B. K., Browne P. J., Lindow S. E. (2010). Novel high throughput detection method to assess bacterial surfactant production. Appl. Environ. Microbiol. 76 5363–5372. 10.1128/aem.00592-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch A. Y., Zeisler V., Yokota K., Schreiber L., Lindow S. E. (2014). The hygroscopic biosurfactant syringafactin produced by Pseudomonas syringae enhances fitness on leaf surfaces during fluctuating humidity. Environ. Microbiol. 16 2086–2098. 10.1111/1462-2920.12437 [DOI] [PubMed] [Google Scholar]
- Burrowes E., Baysse C., Adams C., O’Gara F. (2006). Influence of the regulatory protein RsmA on cellular functions in Pseudomonas aeruginosa PAO1, as revealed by transcriptome analysis. Microbiology 152(Pt 2) 405–418. 10.1099/mic.0.28324-0 [DOI] [PubMed] [Google Scholar]
- Campisano A., Schroeder C., Schemionek M., Overhage J., Rehm B. H. A. (2006). PslD is a secreted protein required for biofilm formation by Pseudomonas aeruginosa. Appl. Environ. Microbiol. 72 3066–3068. 10.1128/aem.72.4.3066-3068.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A., Cui Y., Yang H., Collmer A., Alfano J., Chatterjee A. (2003). GacA, the response regulator of a two-component system, acts as a master regulator in Pseudomonas syringae pv. tomato DC3000 by controlling regulatory RNA, transcriptional activators, and alternate sigma factors. Mol. Plant Microbe Interact. 16 1106–1117. 10.1094/mpmi.2003.16.12.1106 [DOI] [PubMed] [Google Scholar]
- Clarke C. R., Hayes B. W., Runde B. J., Markel E., Swingle B. M., Vinatzer B. A. (2016). Comparative genomics of Pseudomonas syringae pathovar tomato reveals novel chemotaxis pathways associated with motility and plant pathogenicity. Peer J. 4:e2570. 10.7717/peerj.2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y., Chatterjee A., Chatterjee A. K. (2001). Effect of the two-component system comprising GacA and GacS of Erwinia carotovora subsp. carotovora on the production of global regulatory rsmB RNA, extracellular enzymes, and HarpinEcc. Mol. Plant Microbe Interact. 14 516–526. 10.1094/mpmi.2001.14.4.516 [DOI] [PubMed] [Google Scholar]
- Dugar G., Svensson S. L., Bischler T., Wäldchen S., Reinhardt R., Sauer M., et al. (2016). The CsrA-FliW network controls polar localization of the dual-function flagellin mRNA in Campylobacter jejuni. Nat. Commun. 7:11667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duss O., Michel E., Yulikov M., Schubert M., Jeschke G., Allain F. H. T. (2014). Structural basis of the non-coding RNA RsmZ acting as a protein sponge. Nature 509 588–592. 10.1038/nature13271 [DOI] [PubMed] [Google Scholar]
- Ferreiro M. D., Nogales J., Farias G. A., Olmedilla A., Sanjuán J., Gallegos M. T. (2018). Multiple CsrA proteins control key virulence traits in Pseudomonas syringae pv. tomato DC3000. Mol. Plant Microbe Interact. 31 525–536. 10.1094/mpmi-09-17-0232-r [DOI] [PubMed] [Google Scholar]
- Ge Y. X., Lee J. H., Hu B. S., Zhao Y. F. (2018). Loss-of-function mutations in the Dpp and Opp permeases render Erwinia amylovora resistant to kasugamycin and blasticidin S. Mol. Plant Microbe Interact. 31 823–832. 10.1094/mpmi-01-18-0007-r [DOI] [PubMed] [Google Scholar]
- Ge Y. X., Lee J. H., Liu J., Yang H. W., Tian Y. L., Hu B. S., et al. (2019). Homologues of the RNA binding protein RsmA in Pseudomonas syringae pv. tomato DC3000 exhibit distinct binding affinities with non-coding small RNAs and have distinct roles in virulence. Mol. Plant Pathol. 20 1217–1236. 10.1111/mpp.12823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudapaty S., Suzuki K., Wang X., Babitzke P., Romeo T. (2001). Regulatory interactions of Csr components: the RNA binding protein CsrA activates csrB transcription in Escherichia coli. J. Bacteriol. 183 6017–6027. 10.1128/jb.183.20.6017-6027.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman D. S., Vinatzer B. A., Sarkar S. F., Ranall M. V., Kettler G., Greenberg J. T. (2002). A functional screen for the type III (Hrp) secretome of the plant pathogen Pseudomonas syringae. Science 295 1722–1726. 10.1126/science.295.5560.1722 [DOI] [PubMed] [Google Scholar]
- Heeb S., Haas D. (2001). Regulatory roles of the GacS/GacA two component system in plant-associated and other gram-negative bacteria. Mol. Plant Microbe Interact. 14 1351–1363. 10.1094/mpmi.2001.14.12.1351 [DOI] [PubMed] [Google Scholar]
- Hickman J. W., Tifrea D. F., Harwood C. S. (2005). A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc. Natl. Acad. Sci. U.S.A. 102 14422–14427. 10.1073/pnas.0507170102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmqvist E., Wright P. R., Li L., Bischler T., Barquist L., Reinhardt R., et al. (2016). Global RNA recognition patterns of post-transcriptional regulators Hfq and CsrA revealed by UV crosslinking in vivo. EMBO J. 35 991–1011. 10.15252/embj.201593360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas-Rosales Ó, Ramos-González M. I., Espinosa-Urgel M. (2016). Self-regulation and interplay of Rsm family proteins modulate the lifestyle of Pseudomonas putida. Appl. Environ. Microbiol. 82 5673–5686. 10.1128/aem.01724-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh T. V., Dahlbeck D., Staskawicz B. J. (1989). Bacterial blight of soybean: regulation of a pathogen gene determining host cultivar specificity. Science 245 1374–1377. 10.1126/science.2781284 [DOI] [PubMed] [Google Scholar]
- Ishiga T., Ishiga Y., Betsuyaku S., Nomura N. (2018). AlgU contributes to the virulence of Pseudomonas syringae pv. tomato DC3000 by regulating production of the phytotoxin coronatine. J. Gen. Plant Pathol. 84 189–201. 10.1007/s10327-018-0775-6 [DOI] [Google Scholar]
- Ishiga Y., Ichinose Y. (2016). Pseudomonas syringae pv. tomato OxyR is required for virulence in tomato and Arabidopsis. Mol. Plant Microbe Interact. 29 119–131. 10.1094/mpmi-09-15-0204-r [DOI] [PubMed] [Google Scholar]
- Janssen K. H., Diaz M. R., Gode C. J., Wolfgang M. C., Yahr T. L. (2018). RsmV, a small noncoding regulatory RNA in Pseudomonas aeruginosa that sequesters RsmA and RsmF from target mRNAs. J. Bacteriol. 200:e00277-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia D., Merey G., Nakayama S., Zheng Y., Zhou J., Luo Y., et al. (2013). Nucleotide, c-di-GMP, c-di-AMP, cGMP, cAMP, (p)ppGpp signaling in bacteria and implications in pathogenesis. Chem. Soc. Rev. 42 305–341. 10.1039/c2cs35206k [DOI] [PubMed] [Google Scholar]
- Kasapis S., Morris E. R., Gross M., Rudolph K. (1994). Solution properties of levan polysaccharide from Pseudomonas syringae pv. phaseolicola, and its possible role as a blocker of recognition during pathogenesis. Carbohydr. Polym. 23 55–64. 10.1016/0144-8617(94)90090-6 [DOI] [Google Scholar]
- Kay E., Humair B., Dénervaud V., Riedel K., Spahr S., Eberl L., et al. (2006). Two GacA-dependent small RNAs modulate the quorum-sensing response in Pseudomonas aeruginosa. J. Bacteriol. 188 6026–6033. 10.1128/jb.00409-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapouge K., Schubert M., Allain F. H., Haas D. (2008). Gac/Rsm signal transduction pathway of γ-proteobacteria: from RNA recognition to regulation of social behaviour. Mol. Microbiol. 67 241–253. 10.1111/j.1365-2958.2007.06042.x [DOI] [PubMed] [Google Scholar]
- Lawhon S. D., Frye J. G., Suyemoto M., Porwollik S., McClelland M., Altier C. (2003). Global regulation by CsrA in Salmonella typhimurium. Mol. Microbiol. 48 1633–1645. 10.1046/j.1365-2958.2003.03535.x [DOI] [PubMed] [Google Scholar]
- Lee J. H., Ancona V., Chatnaparat T., Zhao Y. F. (2019). The RNA-binding protein CsrA controls virulence in Erwinia amylovora by regulating RelA, RcsB, and FlhD at the posttranscriptional level. Mol. Plant Microbe Interact. 32 1448–1459. 10.1094/mpmi-03-19-0077-r [DOI] [PubMed] [Google Scholar]
- Lindow S. E. (1991). “Determinants of epiphytic fitness in bacteria,” in Microbial Ecology of Leaves, eds Andres J. H., Hirano S. S. (New York, NY: Springer Verlag; ), 295–314. 10.1007/978-1-4612-3168-4_15 [DOI] [Google Scholar]
- Liu J., Yu M. G., Chatnaparat T., Lee J. H., Tian Y. L., Hu B. S., et al. (2020). Comparative transcriptomic analysis of global gene expression mediated by (p) ppGpp reveals common regulatory networks in Pseudomonas syringae. BMC Genom. 21:296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marden J. N., Diaz M. R., Walton W. G., Gode C. J., Betts L., Urbanowski M. L., et al. (2013). An unusual CsrA family member operates in series with RsmA to amplify posttranscriptional responses in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 110 15055–15060. 10.1073/pnas.1307217110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matas I. M., Lambertsen L., Rodríguez-Moreno L., Ramos C. (2012). Identification of novel virulence genes and metabolic pathways required for full fitness of Pseudomonas savastanoi pv. savastanoi in olive (Olea europaea) knots. New Phytologist 196 1182–1196. 10.1111/j.1469-8137.2012.04357.x [DOI] [PubMed] [Google Scholar]
- Miller M. B., Bassler B. L. (2001). Quorum sensing in bacteria. Annu. Rev. Microbiol. 55 165–199. [DOI] [PubMed] [Google Scholar]
- Moll S., Schneider D. J., Stodghill P., Myers C. R., Cartinhour S. W., Filiatrault M. J. (2010). Construction of an rsmX co-variance model and identification of five rsmX non-coding RNAs in Pseudomonas syringae pv. tomato DC3000. RNA Biol. 7 508–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulcahy H., O’Callaghan J., O’Grady E. P., Adams C., O’Gara F. (2006). The posttranscriptional regulator RsmA plays a role in the interaction between Pseudomonas aeruginosa and human airway epithelial cells by positively regulating the type III secretion system. Infect. Immun. 74 3012–3015. 10.1128/iai.74.5.3012-3015.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales J., Vargas P., Farias G. A., Olmedilla A., Sanjuan J., Gallegos M. T. (2015). FleQ coordinates flagellum dependent and independent motilities in Pseudomonas syringae pv. tomato DC3000. Appl. Environ. Microbiol. 81 7533–7545. 10.1128/aem.01798-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overhage J., Schemionek M., Webb J. S., Rehm B. H. A. (2005). Expression of the psl operon in Pseudomonas aeruginosa PAO1 biofilms: PslA performs an essential function in biofilm formation. Appl. Environ. Microbiol. 71 4407–4413. 10.1128/aem.71.8.4407-4413.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penaloza-Vazquez A., Fakhr M. K., Bailey A. M., Bender C. L. (2004). AlgR functions in algC expression and virulence in Pseudomonas syringae pv. syringae. Microbiology 150 2727–2737. 10.1099/mic.0.27199-0 [DOI] [PubMed] [Google Scholar]
- Penaloza-Vazquez A., Kidambi S. P., Chakrabarty A. M., Bender C. L. (1997). Characterization of the alginate biosynthetic gene cluster in Pseudomonas syringae pv. syringae. J. Bacteriol. 179 4464–4472. 10.1128/jb.179.14.4464-4472.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Mendoza D., Aragón I. M., Prada-Ramírez H. A., Romero-Jiménez L., Ramos C., Gallegos M. T., et al. (2014). Responses to elevated c-di-GMP levels in mutualistic and pathogenic plant-interacting bacteria. PLoS One 9:e91645. 10.1371/journal.pone.0091645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessi G., Williams F., Hindle Z., Heurlier K., Holden M. T., Camara M., et al. (2001). The global posttranscriptional regulator RsmA modulates production of virulence determinants and N-acylhomoserine lactones in Pseudomonas aeruginosa. J. Bacteriol. 183 6676–6683. 10.1128/jb.183.22.6676-6683.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts A. H., Vakulskas C. A., Pannuri A., Yakhnin H., Babitzke P., Romeo T. (2017). Global role of the bacterial post-transcriptional regulator CsrA revealed by integrated transcriptomics. Nat. Commun. 8:1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourciau C., Lai Y. J., Gorelik M., Babitzke P., Romeo T. (2020). Diverse mechanisms and circuitry for global regulation by the RNA-binding Protein CsrA. Front. Microbiol. 27:601352. 10.3389/fmicb.2020.601352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prada-Ramírez H. A., Pérez-Mendoza D., Felipe A., Martínez-Granero F., Rivilla R., Sanjuán J., et al. (2016). AmrZ regulates cellulose production in Pseudomonas syringae pv. tomato DC3000. Mol. Microbiol. 99 960–977. [DOI] [PubMed] [Google Scholar]
- Ramírez-Zapata D., Ramos C., Aguilera S., Bardaji L., Martínez-Gil M., Murillo J. (2020). Two homologues of the global regulator Csr/Rsm redundantly control phaseolotoxin biosynthesis and virulence in the plant pathogen Pseudomonas amygdali pv. phaseolicola 1448A. Microorganisms 8:1536. 10.3390/microorganisms8101536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey D. M., Wozniak D. J. (2005). Understanding the control of Pseudomonas aeruginosa alginate synthesis and the prospects for management of chronic infections in cystic fibrosis. Mol. Microbiol. 56 309–322. 10.1111/j.1365-2958.2005.04552.x [DOI] [PubMed] [Google Scholar]
- Reimmann C., Valverde C., Kay E., Haas D. (2005). Posttranscriptional repression of GacS/GacA-controlled genes by the RNA-binding protein RsmE acting together with RsmA in biocontrol strain Pseudomonas fluorescens CHA0. J. Bacteriol. 187 276–285. 10.1128/jb.187.1.276-285.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J., Smyth G. K. (2010). EdgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo T., Vakulsas C. A., Babitzke P. (2013). Post-transcriptional regulation on a global scale: form and function of Csr/Rsm systems. Environ. Microbiol. 15 313–324. 10.1111/j.1462-2920.2012.02794.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahr T., Rusniok C., Impens F., Oliva G., Sismeiro O., Coppée J. Y., et al. (2017). The Legionella pneumophila genome evolved to accommodate multiple regulatory mechanisms controlled by the CsrA-system. PLoS Genet. 13:e1006629. 10.1371/journal.pgen.1006629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schechter L. M., Roberts K. A., Jamir Y., Alfano J. R., Collmer A. (2004). Pseudomonas syringae type III secretion system targeting signals and novel effectors studied with a Cya translocation reporter. J. Bacteriol. 186 543–555. 10.1128/jb.186.2.543-555.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert M., Lapouge K., Duss O., Oberstrass F. C., Jelesarov I., Haas D., et al. (2007). Molecular basis of messenger RNA recognition by the specific bacterial repressing clamp RsmA/CsrA. Nat. Struct. Mol. Biol. 14 807–813. 10.1038/nsmb1285 [DOI] [PubMed] [Google Scholar]
- Sobrero P. M., Valverde C. (2020). Comparative genomics and evolutionary analysis of RNA-binding proteins of the CsrA family in the genus Pseudomonas. Front. Mol. Biosci. 7:127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnleitner E., Abdou L., Haas D. (2009). Small RNA as global regulator of carbon catabolite repression in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 106 21866–21871. 10.1073/pnas.0910308106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreedharan A., Penaloza-Vazquez A., Kunkel B. N., Bender C. L. (2006). CorR regulates multiple components of virulence in Pseudomonas syringae pv. tomato DC3000. Mol. Plant Microbe Interact. 19 768–779. 10.1094/mpmi-19-0768 [DOI] [PubMed] [Google Scholar]
- Stock A. M., Robinson V. L., Goudreau P. N. (2000). Two-component signal transduction. Annu. Rev. Biochem. 69 183–215. [DOI] [PubMed] [Google Scholar]
- Suzuki K., Wang X., Weilbacher T., Pernestig A. K., Melefors O., Georgellis D., et al. (2002). Regulatory circuitry of the CsrA/CsrB and BarA/UvrY systems of Escherichia coli. J. Bacteriol. 184 5130–5140. 10.1128/jb.184.18.5130-5140.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Xiao Y., Zhou J. M. (2006). Regulation of the type III secretion system in phytopathogenic bacteria. Mol. Plant Microbe Interact. 19 1159–1166. 10.1094/mpmi-19-1159 [DOI] [PubMed] [Google Scholar]
- Vakulskas C. A., Potts A. H., Babitzke P., Ahmer B. M., Romeo T. (2015). Regulation of bacterial virulence by Csr (Rsm) systems. Microbiol. Mol. Biol. Rev. 79 193–224. 10.1128/mmbr.00052-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadhams G. H., Armitage J. P. (2004). Making sense of it all: bacterial chemotaxis. Nat. Rev. Mol. Cell Biol. 5 1024–1037. 10.1038/nrm1524 [DOI] [PubMed] [Google Scholar]
- Wang X., Alarcón-Chaidez F., Peñaloza-Vázquez A., Bender C. L. (2002). Differential regulation of coronatine biosynthesis in Pseudomonas syringae pv. tomato DC3000 and P. syringae pv. glycinea PG4180. Physiol. Mol. Plant Pathol. 60 111–120. 10.1006/pmpp.2002.0382 [DOI] [Google Scholar]
- Wei B. L., Brun-Zinkernagel A. M., Simecka J. W., Prüß B. M., Babitzke P., Romeo T. (2001). Positive regulation of motility and flhDC expression by the RNA−binding protein CsrA of Escherichia coli. Mol. Microbiol. 40 245–256. 10.1046/j.1365-2958.2001.02380.x [DOI] [PubMed] [Google Scholar]
- Whalen M., Innes R., Bent A., Staskawicz B. (1991). Identification of Pseudomonas syringae pathogens of Arabidopsis thaliana and a bacterial gene determining avirulence on both Arabidopsis and soybean. Plant Cell 3 49–59. 10.1105/tpc.3.1.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Shao X., Deng X. (2019). Regulation of type III secretion system in Pseudomonas syringae. Environ. Microbiol. 21 4465–4477. 10.1111/1462-2920.14779 [DOI] [PubMed] [Google Scholar]
- Xin X. F., He S. Y. (2013). Pseudomonas syringae pv. tomato DC3000: a model pathogen for probing disease susceptibility and hormone signaling in plants. Annu. Rev. Phytopathol. 51 473–498. 10.1146/annurev-phyto-082712-102321 [DOI] [PubMed] [Google Scholar]
- Yakhnin A. V., Baker C. S., Vakulskas C. A., Yakhnin H., Berezin I., Romeo T., et al. (2013). CsrA activates flhDC expression by protecting flhDC mRNA from RNase E-mediated cleavage. Mol. Microbiol. 87 851–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Thilmony R., Bender C. L., Schaller A., He S. Y., Howe G. A. (2003). Virulence systems of Pseudomonas syringae pv. tomato promote bacterial speck disease in tomato by targeting the jasmonate signaling pathway. Plant J. 36 485–499. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multidimensional clustering plot. Dimension 1 and Dimension 2 explained data variability of 60% (media) and 21% (strains), respectively, for strains of PstDC3000, ΔrsmA3, and ΔrsmA23 cultured in HMM and KB media. HMM: hrp-inducing minimum medium; KB: King’s B medium; PstDC3000: Pseudomonas syringae pv. tomato DC3000; ΔrsmA3: the rsmA3 mutant; ΔrsmA23: the rsmA2/A3 double mutant.
Heatmap. (A) PstDC3000, ΔrsmA3, and ΔrsmA23 in both HMM and KB, with a total of 2661 differentially expressed genes (DEGs). (B) PstDC3000, ΔrsmA3, and ΔrsmA23 in HMM with a total of 1560 DEGs. (C) PstDC3000, rsmA3, and rsmA23 in KB with a total of 1879 DEGs. Up- and down-regulated genes were indicated by red and blue colors, respectively. HMM: hrp-inducing minimum medium; KB: King’s B medium; PstDC3000: Pseudomonas syringae pv. tomato DC3000; ΔrsmA3: the rsmA3 mutant; ΔrsmA23: the rsmA2/A3 double mutant.
MA plots (a.k.a., mean-difference plots; M: log2FC; A: average log2CPM; CPM: counts per million reads). (A) ΔrsmA23 versus PstDC3000 in HMM. (B) ΔrsmA23 versus ΔrsmA3 in HMM. (C) ΔrsmA3 versus PstDC3000 in HMM. (D) ΔrsmA23 versus PstDC3000 in KB. (E) ΔrsmA23 versus ΔrsmA3 in KB. (F) ΔrsmA3 versus PstDC3000 in KB. M: log2FC, A: Average Log2CPM, counts per million reads. Dots between two purple lines represent | log2FC| value ≤ 0.58496250072 (i.e., | FC| value ≤ 1.5), and outside dots represent | log2FC| value ≥ 0.58496250072 (i.e., | FC| value ≥ 1.5). Up- and down-regulated genes were indicated by red and blue dot with p value < 0.05, black dot represents no significant difference with p value > 0.05. HMM: hrp-inducing minimum medium; KB: King’s B medium; PstDC3000: Pseudomonas syringae pv. tomato DC3000; ΔrsmA3: the rsmA3 mutant; ΔrsmA23: the rsmA2/A3 double mutant.
Functional classification of differentially expressed genes (DEGs) based on the clusters of orthologous groups (COGs). (A) ΔrsmA23 versus PstDC3000 in HMM. (B) ΔrsmA23 versus ΔrsmA3 in HMM. (C) ΔrsmA3 versus PstDC3000 in HMM. (D) ΔrsmA23 versus PstDC3000 in KB. (E) ΔrsmA23 versus ΔrsmA3 in KB. (F) ΔrsmA3 versus PstDC3000 in KB. DEGs were defined as genes with a | Fold Change (FC)| value ≥ 1.5 and a p value < 0.05. Up- and down-regulated DEGs were indicated by red and blue bars, respectively. HMM: hrp-inducing minimum medium; KB: King’s B medium; PstDC3000: Pseudomonas syringae pv. tomato DC3000; ΔrsmA3: the rsmA3 mutant; ΔrsmA23: the rsmA2/A3 double mutant.
Verification of selected differentially expressed genes (DEGs) using quantitative real-time PCR. (A) Expression of the katE, cyaA, fleQ, phoB, citM, ladS, and pyk genes in the rsmA3 mutant and the rsmA2/A3 double mutant as compared with PstDC3000 grown in HMM. (B) Expression of the katE, cyaA, fleQ, phoB, citM, ladS, and pyk genes in the rsmA3 mutant and the rsmA2/A3 double mutant as compared with PstDC3000 in KB. The values on top of bars represent fold changes of the gene in ΔrsmA23 or ΔrsmA3 versus PstDC3000, while the absence of a value represents no significant difference in the RNA-seq data. HMM: hrp-inducing minimum medium; KB: King’s B medium; PstDC3000: Pseudomonas syringae pv. tomato DC3000; ΔrsmA3: the rsmA3 mutant; ΔrsmA23: the rsmA2/A3 double mutant.
Primers for qRT-PCR used in this study.
List of differentially expressed genes (DEGs) in PstDC3000, the rsmA3 mutant, and the rsmA2/A3 double mutant in HMM.
List of differentially expressed genes (DEGs) in PstDC3000, the rsmA3 mutant, and the rsmA2/A3 double mutant in KB.
Fold changes of the type III secretion system (T3SS) and alginate biosynthesis genes in HMM.
Fold changes of the syringafactin biosynthesis genes in HMM.
Selected genes regulated mainly by RsmA2 in HMM.
Selected genes regulated mainly by RsmA3 in HMM.
Fold changes of the chemotaxis genes in KB.
Fold changes of the ribosome genes in KB.
Selected genes regulated mainly by RsmA2 in KB.
Selected genes regulated mainly by RsmA3 in KB.
Data Availability Statement
RNA-seq data files have been submitted to Gene Expression Omnibus (GEO) at the National Center for Biotechnology Information (NCBI) with an accession number GSE162091.