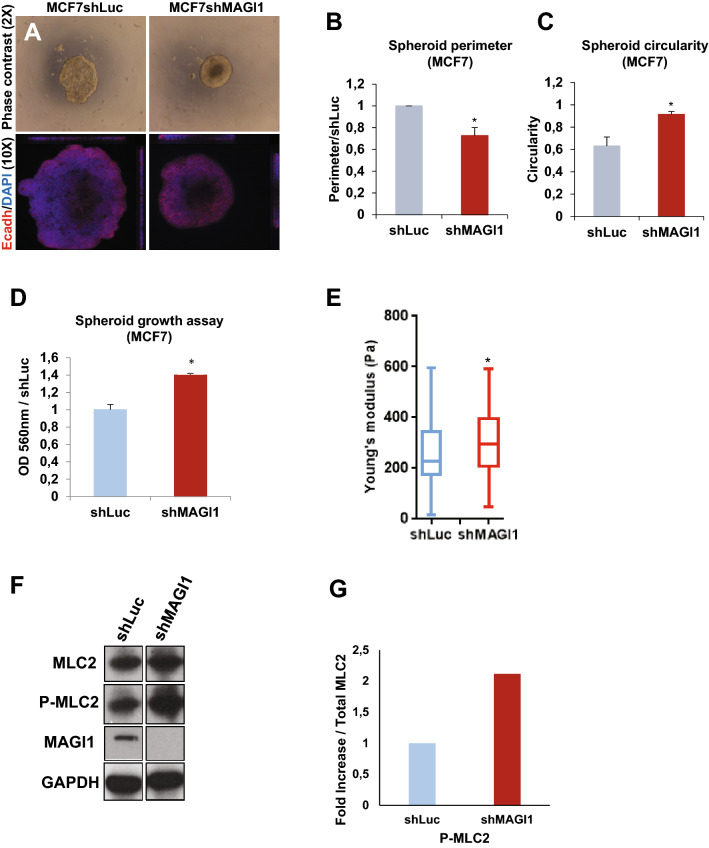
Figure 4.
The loss of MAGI1 affects MCF7 cell compaction, ROCK activity, and compressive forces. (A) Representative phase contrast and fluorescence images of MCF7shLuc and MCF7shMAGI1 cells grown in 3D spheroid cultures and stained with E-cadherin (red) and DAPI (blue). (B) Calculated perimeters for 3D spheroid cultures of MCF7shMAGI1 normalized by the perimeter of MCF7shLuc cells. Bars represent mean ± SD (n = 10 spheroids) of five independent experiments. Unpaired two-tailed Student’s t-test; *p < 0.05. (C) Calculated circularity for 3D spheroid cultures of MCF7shLuc and MCF7shMAGI1 (calculations were done with the ImageJ software where a value of 1 is considered as a perfect circle). Bars represent mean ± SD (n = 10 spheroids) of five independent experiments. Unpaired two-tailed Student’s t-test; *p < 0.05. (D) MTT assay (OD 560 nm) representing 3D spheroid cell growth of MCF7shMAGI1 compared to MCF7shLuc cells 5 days after cell seeding. Bars represent mean ± SD (n = 10 spheroids) of five independent experiments. Unpaired two-tailed Student’s t-test; *p < 0.05. (E) Elastic Young’s modulus (EYM) of cells: Hertz contact mechanics model for spherical indenters was used. In MCF7shMAGI1 cells, the apical surface EYM is significantly elevated when compared to control MCF7shLuc cells. Data are represented as mean + /− SD: MCF7shLuc EYM = 258.8 + /− 101, MCF7shMAGI1 EYM = 302.4 + /− 91. Unpaired two-tailed Student’s t-test; *p < 0.05 (n = 132 and 94 for MCF7shLuc and MCF7shMAGI1 respectively). (F) Western blot analysis on whole protein extracts (n = 3) of ROCK-specific ser19 phosphorylation of Myosin Light Chain 2, total MLC2 and MAGI1 in MCF7shMAGI1 cell lines compared to MCF7shLuc. GAPDH was used as a loading control. Uncropped blots can be found in the Supplementary Information. (G) Quantification of the representative Western blot showing protein expression represented in panel E. Phosphorylated proteins were quantified as compared to their total protein counterparts to evaluate their activation.