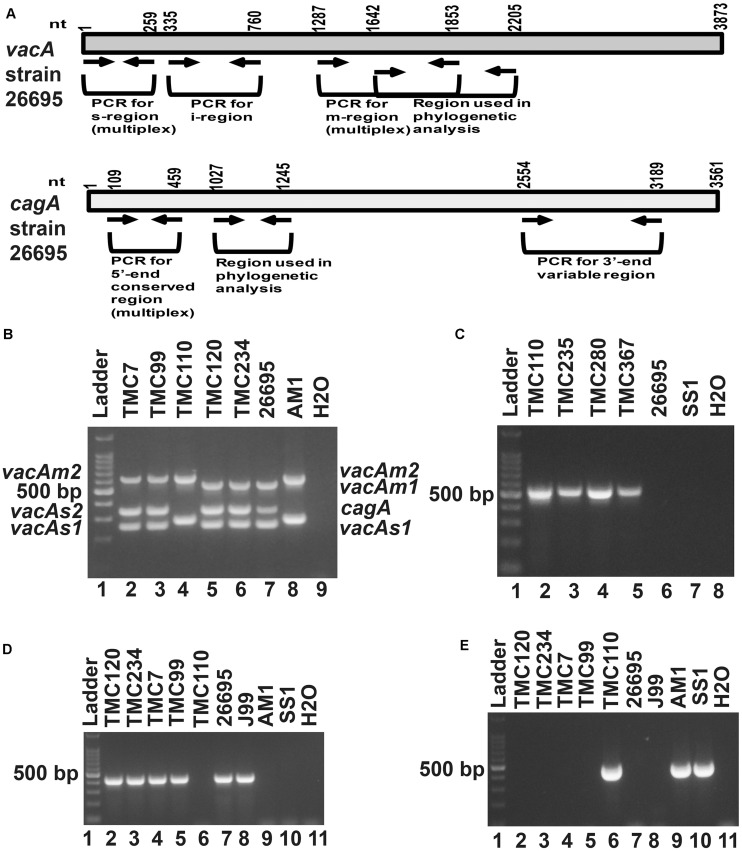
FIGURE 1.
(A) Schematic diagram of the vacuolating cytotoxin A (vacA) and cytotoxin associated gene A (cagA) of the genome sequenced strain, 26,695 showing the nucleotide positions. The regions used in the analyses are indicated. (B) Multiplex PCR for genotyping the vacAs1, vacAs2, vacAm1, vacAm2, and cagA of H. pylori. The strains TMC7 and TMC99 (lane 2 and 3) have vacAs1m2cagA+; the strain TMC110 (lane 4) has vacAs2m2cagA–; the strains TMC120 and TMC234 (Lanes 5 and 6) have vacAs1m1cagA+ genotypes. The strains 26,695 (lane 7) and AM1 (lane 8) were used as positive controls for vacAs1m1cagA+ and vacAs2m2cagA- genotypes, respectively. Water (lane 9) was used as negative control. (C) The strains, TMC110, TMC235, TMC280, TMC367, which did not give amplicon for cagA in multiplex PCR, gave amplicon for the cag-empty site PCR. (D) PCR for the detection of vacAi1 allele. The strains TMC120, TMC 234, TMC7, and TMC99 were found to carry the vacAi1 allele, while the strain TMC110 did not give the amplicon. (E) PCR for the detection of vacAi2 allele. The strain TMC110 was found to carry the vacAi2 allele.