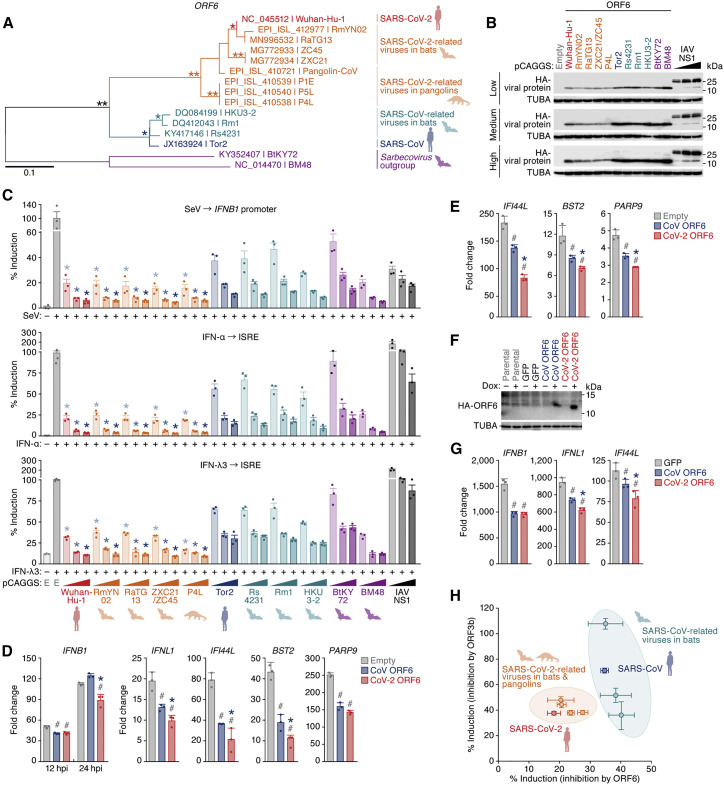
Figure 2.
Potent anti-innate immune signaling activity of Sarbecovirus ORF6
(A) A maximum likelihood phylogenetic tree of Sarbecovirus ORF6 sequences. Accession number, strain name, and the host of each virus are indicated. A scale bar indicates 0.1 nucleotide substitutions per site. Bootstrap values; ∗∗, > 95%; ∗, > 75%.
(B and C) Potent anti-IFN activity of Sarbecovirus ORF6. HEK293 cells were cotransfected with three different amounts of plasmids expressing hemagglutinin (HA)-tagged Sarbecovirus ORF6 or IAV NS1 and p125Luc (C, top) or pISRE-luc (C, center and bottom). 24 h after transfection, cells were infected with SeV (MOI 10) (C, top) or treated with IFN-α (C, center) or IFN-λ3 (C, bottom). 24 h after infection or treatment, cells were harvested for western blotting (B) and a luciferase assay (C). Note that the ORF6 sequence of ZXC21 is identical to that of ZC45. In (B), “low,” “medium,” and “high” indicate the amount of transfected Sarbecovirus ORF6 expression plasmids.
(D and E) Anti-innate immune signaling activity of SARS-CoV-2 ORF6 in HEK293 cells. HEK293 cells were transfected with plasmids expressing HA-tagged ORF6. 24 h after transfection, cells were infected with SeV (MOI 10) (D) or treated with IFN-α (E). 12 h (IFNB1 only) or 24 h (all genes) after infection (D) or 8 h after IFN-α treatment (E), endogenous expression levels of the indicated genes were measured by real-time RT-PCR.
(F and G) Anti-IFN activity of SARS-CoV-2 ORF6 in lung cells.
(F) Expression of HA-tagged ORF6 upon Dox stimulation.
(G) A549 cells stably transduced with a Dox-inducible HA-tagged ORF6 or GFP (F) were treated with Dox and infected with SeV (MOI 10). 4 h after infection, the endogenous expression levels of the indicated genes were measured by real-time RT-PCR.
(H) Comparison of the anti-IFN activity of Sarbecovirus ORF6 and ORF3b. The anti-IFN activities of ORF6 (100 ng, x axis) and ORF3b (100 ng, y axis) (Figures S1C and S1D) are summarized.
For western blotting (B and F), the input of the cell lysate was normalized to TUBA, and one representative result of three independent experiments is shown. In (B), three different doses of IAV NS1 were used as controls on each membrane. kDa, kilodalton. For the luciferase assay (C), the value was normalized to the unstimulated, empty vector-transfected cells (no SeV infection in the top panel and no IFN treatment in the center and bottom panels). For real-time RT-PCR (D, E, and G), expression of the target gene was normalized to GAPDH, and the fold change to the value of 0 h is shown. For the luciferase assay (C) and real-time RT-PCR (D, E, and G), mean values of three independent experiments with SEM are shown. In (C), statistically significant differences (p < 0.05) compared with the same amount of SARS-CoV (Tor2) ORF6-transfected cells (∗) are shown. In (D), (E), and (G), statistically significant differences (p < 0.05) compared with empty vector-transfected cells (#, D and E), GFP-expressing cells upon Dox stimulation (#, G), and SARS-CoV (Tor2) ORF6-transfected cells (∗) are shown. E, empty vector.
See also Figure S1.