Figure 2.
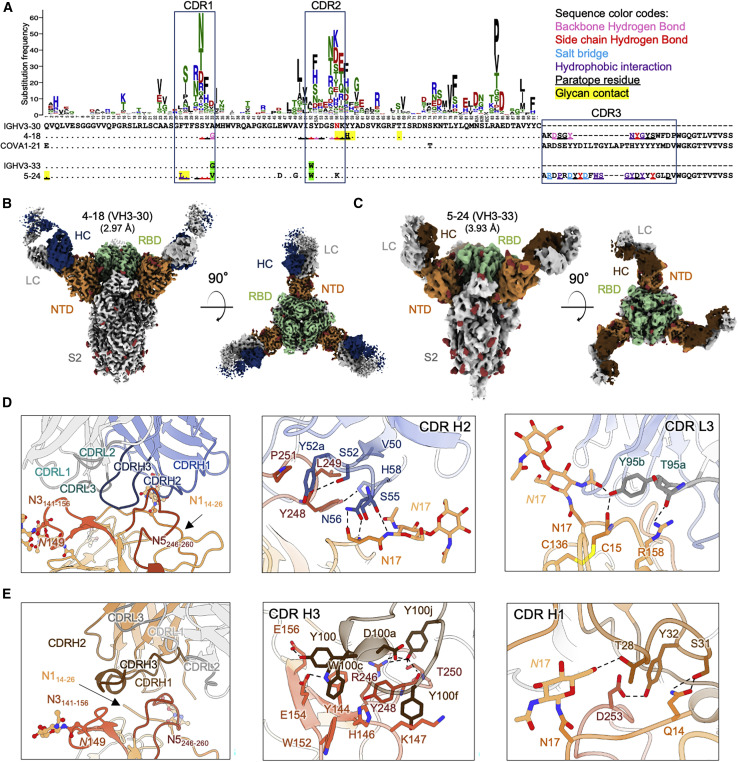
NTD-directed neutralizing antibodies derived from the closely related VH3-30 and VH3-33 genes show distinct binding modes
(A) Sequence alignment of VH3-30-derived (4-18) and VH3-33-derived (5-24) NTD-directed antibodies showing paratope residues, somatic hypermutations, and gene-specific substitution profile (GSSP) showing positional somatic hypermutation probabilities for VH3-30 gene. Substitutions between VH3-30 and VH3-33 germline genes are highlighted in green.
(B) Cryo-EM reconstruction for spike complex with antibody 4-18 from two orthogonal views; NTD is shown in orange, RBD in green, and glycans in red, with antibody heavy chain in blue and light chain in gray.
(C) Cryo-EM reconstruction for spike complex with antibody 5-24 from two orthogonal views; NTD is shown in orange, RBD in green, and glycans in red, with antibody heavy chain in brown and light chain in gray.
(D) Expanded view of 4-18 interactions with NTD showing the overall interface (left), recognition in CDR H2 (middle), and recognition in CDR L3 (right). NTD regions N1 (residues 14–26), N3 (residues 141–156), and N5 (residues 246–260) are shown in shades of orange; CDR H1, H2, and H3 are shown in shades of blue; CDR L1, L2, and L3 are shown in shades of gray.
(E) Expanded view of 5-24 interactions with NTD showing the overall interface (left), recognition in CDR H3 (middle), and recognition in CDR H1 (right), colored as in (D) except for CDR H1, H2, and H3, which are colored in shades of brown.
See also Figures S1, S3, and S6, and Table S1.