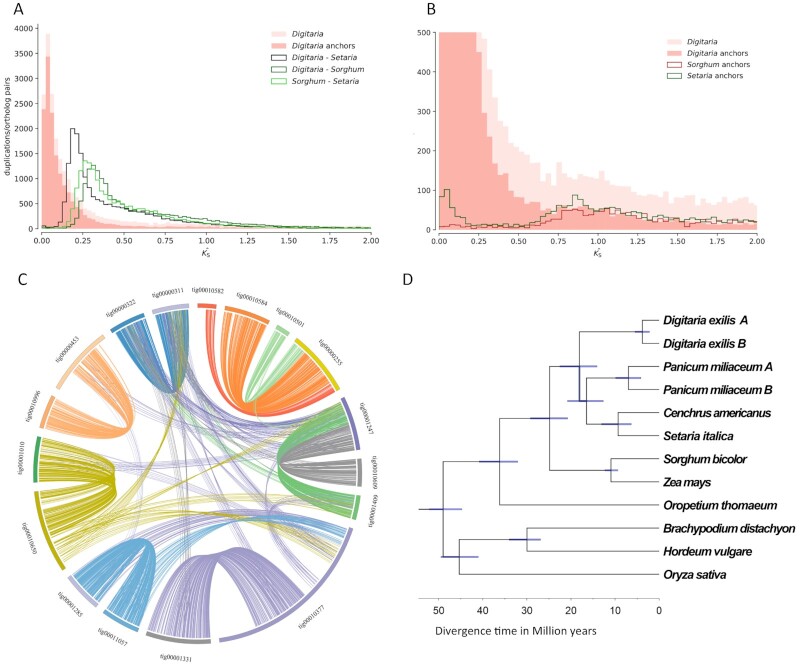
Figure 1:
Whole-genome duplication and polyploidy analysis. (A) Ks estimation of age distribution for paralogs and orthologs of white fonio (Digitaria) and some close relatives. The distribution in light pink represents the entire white fonio paranome, while the distribution in darker pink represents the anchor points (duplicated genes lying in syntenic or collinear regions; see C). Distributions in black, dark green, and light green represent the 1-vs-1 ortholog comparisons between Digitaria-Setaria, Digitaria-Sorghum, and Sorghum-Setaria, respectively. (B) Ks distributions for paralogs of white fonio, sorghum, and Setaria (zoom in), showing an older, likely Poaceae-shared, WGD. (C) Syntenic relationships between putative homoeologous contigs, with colored lines connecting homoeologous gene pairs in the white fonio genome assembly. (D) Time-calibrated phylogenetic tree of several major Poaceae lineages, including white fonio, based on 1,242 gene families consisting of a single gene copy in each lineage and an anchor pair (A and B) in Digitaria. The time scale is shown in million years (My). See text for details.