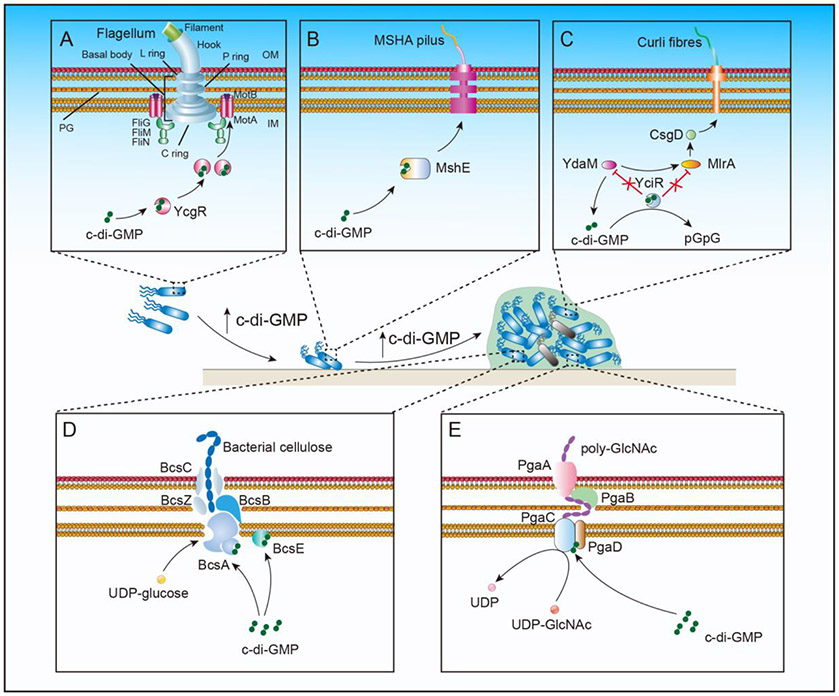
Figure 3. Cyclic di-GMP regulates biofilm formation by inhibiting bacterial motility and increasing EPS production.
(A) Binding of c-di-GMP to the bacterial flagellar brake protein YcgR inhibits the rotation of the flagellar motor, reduces cell motility, and promotes bacterial attachment to the solid surface. (B) Binding of c-di-GMP to MshE promotes the assembly of mannose-sensitive haemagglutinin pilus and helps the bacterial attachment to the solid surface. (C-E) C-di-GMP promotes the synthesis of bacterial EPS and solidifies biofilm formation: (C) When intracellular c-di-GMP concentration reaches a certain threshold, the inhibition of YdaM and MlrA proteins by YciR is relieved. YdaM can activate MlrA to interact with the central curli regulator CsgD, which induces the transcription of curli genes and facilitates curli formation; (D) Bacterial cellulose synthetase catalytic subunit BcsA is anchored on the inner membrane. When c-di-GMP binds to BcsA, its glycosyltransferase domain is activated to assemble the nascent polysaccharide with the help of BcsB/BcsC/BcsZ complex to form extracellular cellulose; (E) The PgaABCD complex promotes formation of the exopolysaccharide poly-GlcNAc. PgaC and PgaD interact with the help of c-di-GMP to form the PgaCD glycosyltransferase complex, which is used for the polymerization and extension of poly-GlcNAc. PgaA and PgaB are responsible for the transport of poly-GlcNAc outside the cell. IM, inner or plasma membrane; PG, peptidoglycan; OM, outer membrane.