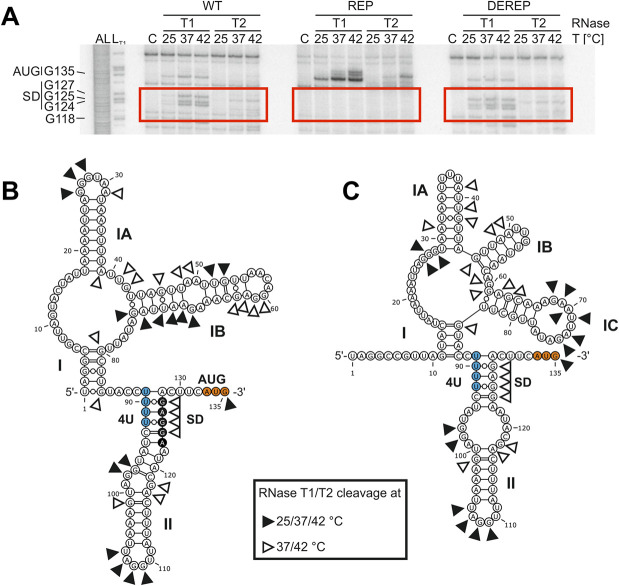
Fig 3. The tviA 5’ UTR fourU RNA thermosensor facilitates temperature-dependent access to the SD sequence.
A) 5’ end-labeled in vitro transcribed RNA containing the wildtype tviA 5’ UTR (WT), the rep3 (T90,92C) tviA 5’ UTR mutant (REP), or the derep4 (T89,91G;C93G) tviA 5’ UTR mutant (DEREP) was enzymatically probed with RNases T1 (cuts 3’ of single-stranded guanines) and T2 (cuts 3’ of single-stranded nucleotides with preference order: A > C > U > G) at 25, 37, and 42°C. Fragmented RNA was separated on an 8% polyacrylamide gel, a portion of which is shown here. AL: alkaline ladder. LT1: RNase T1 cleavage in sequence buffer at 37°C. C: RNA treated with water instead of RNase at 42°C. B) Secondary structure of the tviA 5’ UTR predicted by Mfold [36]. Cleavage sites of RNase T1 and T2, the fourU RNA thermosensor (4U), the Shine-Dalgarno (SD) sequence, and the start codon (AUG) are indicated. C) Secondary structure of the tviA 5’ UTR predicted by Mfold using cleavage information as constraints for folding. Secondary structures were visualized using VARNA applet 3.93 [45]. Data shown in A are representative of 4 independent experiments. See also S4 Fig.