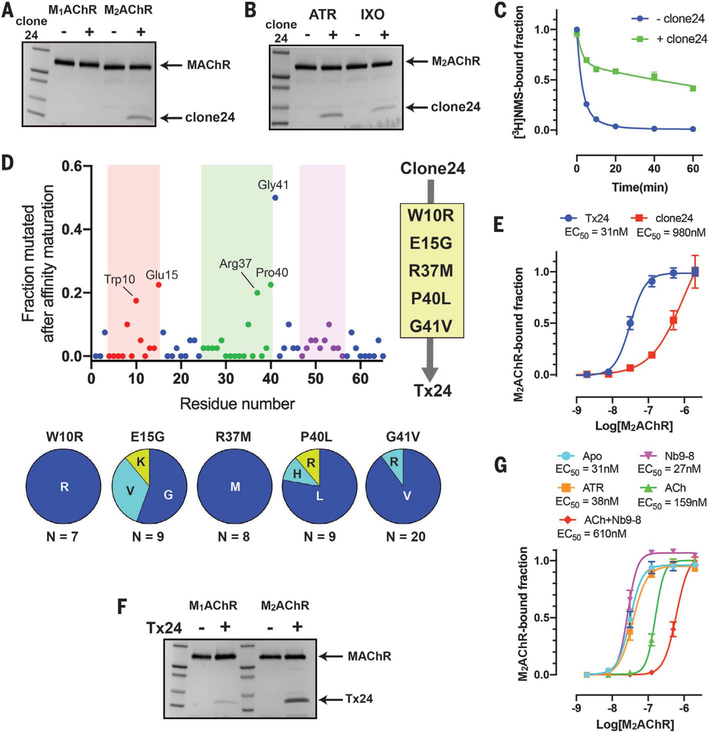
Fig. 3. Identification and affinity maturation of a subtype-selective M2AChR allosteric modulator through in vitro selection.
(A) Pull-down assay showing that clone 24 binds subtype selectively to M2AChR over M1AChR. The experiment was carried out in the presence of 10 μM atropine. (B) Pull-down assay showing the conformation preference of clone 24 toward the atropine (ATR)–bound state over the iperoxo (IXO)–bound state of M2AChR. (C) Comparison of the dissociation kinetics of the orthosteric antagonist [3H]NMS from M2AChR in the clone 24–bound and apo state. Data represent means ± SEM of three experiments performed in triplicate. (D) Sequence analysis of 50 randomly selected clones after the second FACS enrichment. The finger loop regions 1, 2, and 3 are colored red, green, and purple, respectively. The five most frequent mutations (W10R, E15G, R37M, P40L, and G41V) were combined into clone 24 to generate Tx24. (E) Comparison of the affinities of clone 24 and Tx24 to M2AChR using on-yeast affinity measurement in the presence of 10 μM atropine. Data represent means ± SEM of three independent experiments. (F) Pull-down assay of Tx24 using M1AChR or M2AChR in the presence of 10μM atropine. Tx24 prefers M2AChR over M1AChR. (G) Comparison of the affinity of Tx24 for M2AChR bound to different ligands alone or in combination with a G protein mimetic nanobody using on-yeast affinity measurement. Symbols and error bars represent means and SEM of three independent experiments.