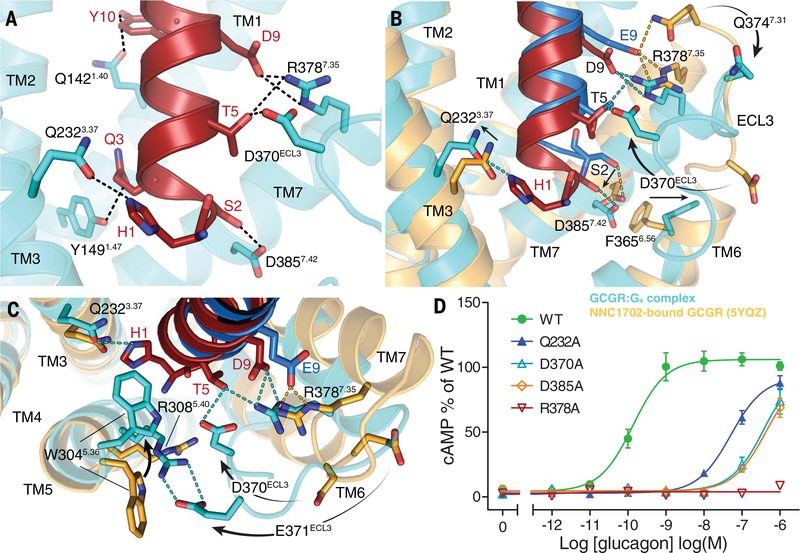
Fig. 2. ZP3780 binding and activation at GCGR.
(A) The N terminus of ZP3780 (red) is required for full ligand efficacy and penetrates deep in the receptor core to make H-bonds (dotted lines) with residues in TM1, TM3, TM7, and ECL3 (cyan). (B) The difference in receptor recognition by full-agonist ZP3780 (red) and partial-agonist NNC1702 (dark blue) that lacks H1 and has a D9E mutation is shown. NNC1702-bound GCGR is shown in orange (PDB 5YQZ) (17), and the ZP3780-bound GCGR-Gs complex structure is shown in cyan. The polar interactions are shown as dotted lines colored according to the GCGR structures bound to the two ligands, respectively (ZP3780, cyan; NNC1702, orange). (C) Comparison of the structures of the partial agonist NNC1702 (blue)–bound GCGR (orange, PDB 5YQZ) (17) and the full-agonist ZP3780 (red)–bound GCGR (cyan) reveal conformational differences in ECL3 and TM5, TM6, and TM7. The presence of H1 in ZP3780 ensures interaction with Q2323.37 and may induce rearrangement of residues in TM5. The interaction of D9 seems to stabilize TM7 and ECL3 displacement, which might trigger GCGR activation. (D) Mutation of Q2323.37, D370ECL3, R3787.35, and D3857.42 to alanine has a large effect on GCGR-mediated cAMP signaling. All mutants were expressed to similar levels as that of the WT receptor. For (D), data represent mean ± SEM from at least three independent experiments, performed in triplicates. Superscripts are Wootten numbering.