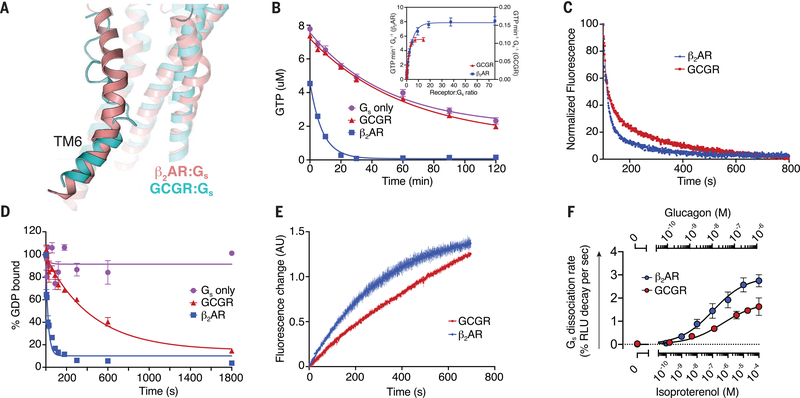
Fig. 5. Engagement and activation of Gs by GCGR and β2AR.
(A) Conformation of TM6 in the GCGR-Gs (cyan) and β2AR-Gs (pink, PDB 3SN6) (50) complex structures. (B) The GTP-turnover assay shows that β2AR (blue) activates Gs much faster than GCGR (red), with GCGR inducing a maximal GTP-turnover rate of 0.11 GTP min−1 Gs−1 compared with (inset) 8 GTP min−1Gs−1 for β2AR. (C) Monitoring of GPCR-Gs association by means of FRET between Cy3B-labeled GCGR (red) or Cy3B-labeled β2AR (blue) with Sulfo-Cy5-labeled Gs. The decrease in donor fluorescence shows comparable rates of association between the receptors and Gs. (D) The rate of receptor-induced [3H]-GDP dissociation from Gs shows faster release for β2AR (blue; koff = 0.042 s−1) compared with GCGR (red; koff = 0.0022 s−1). (E) Bodipy-GTPγS binding to nucleotide-free complex is slower for the GCGR-Gs complex (kon = 0.001 s−1) compared with the β2AR-Gs complex (kon = 0.003 s−1). (F) Concentration-response curves for Gs dissociation rates in HEK293 cells show slower Gs activation by GCGR compared with β2AR. In (B) and (D), data represent mean ± SEM from at least three independent experiments performed in triplicates. In (C) and (E), data represent mean ± SEM of triplicate measurements. In (F), data represent mean ± SEM of three to seven independent experiments.