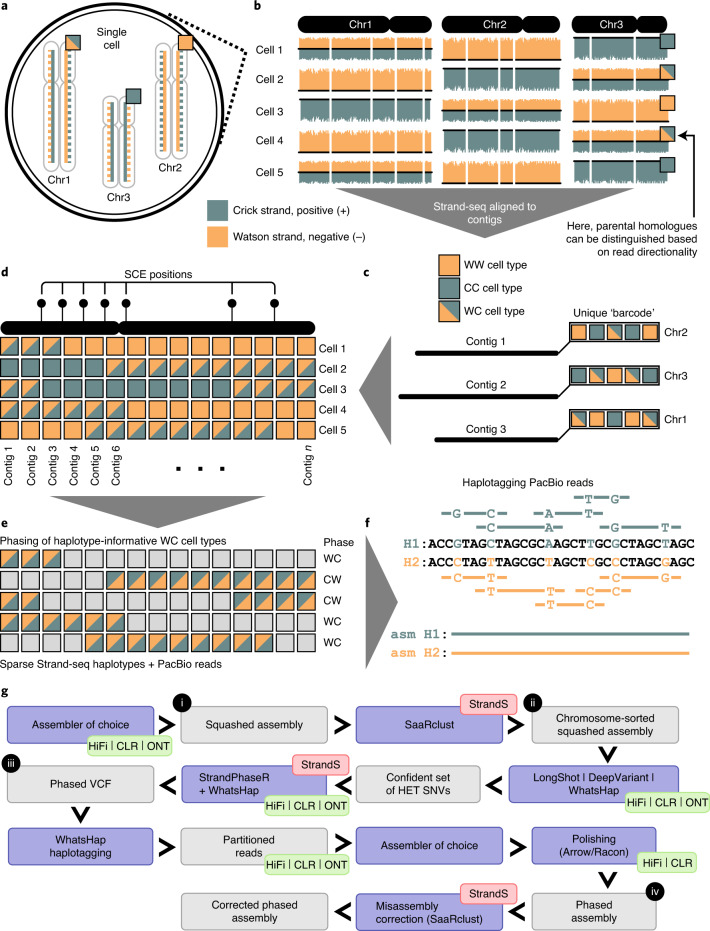
Fig. 1. Overview of the genome assembly pipeline.
a, In a single Strand-seq library, only the template DNA strand (solid line) is sequenced for each parental homologous chromosome. b, Template strands of each homologue in a given diploid cell are randomly inherited by daughter cells (‘+’ positive strand, teal—Crick and ‘−’ negative strand, orange—Watson), resulting in three possible template strand states for homologous chromosomes (height of bars plotted along each chromosome represents the number of ‘+’ and ‘−’ reads mapped in each genomic bin): WC, one Crick and one Watson strand represented for given homologues; WW, only Watson template strands represented; or CC, only Crick template strands represented. c, Unassigned contigs follow the same pattern of template strand state inheritance based on the homologue they belong to. d, Contig order can be inferred based on low-frequency changes in a template strand state resulting from sister chromatid exchange (SCE) events in the parental cell: contigs that are closer to each other tend to share the same template strand state more often than more distant contigs. e, Regions with WC strand state are haplotype informative and can be assembled into continuous haplotypes. f, Haplotypes can then be used to split long reads into their respective homologues. g, Generation of long-read (HiFi/CLR/ONT)-based assemblies: 1) producing squashed assemblies; 2) assigning contigs to clusters using Strand-seq (StrandS); 3) phasing clustered assemblies using the combination of Strand-seq and long PacBio reads; and 4) partitioning and reassembling of haplotype-specific PacBio reads and polishing of the final diploid assemblies.