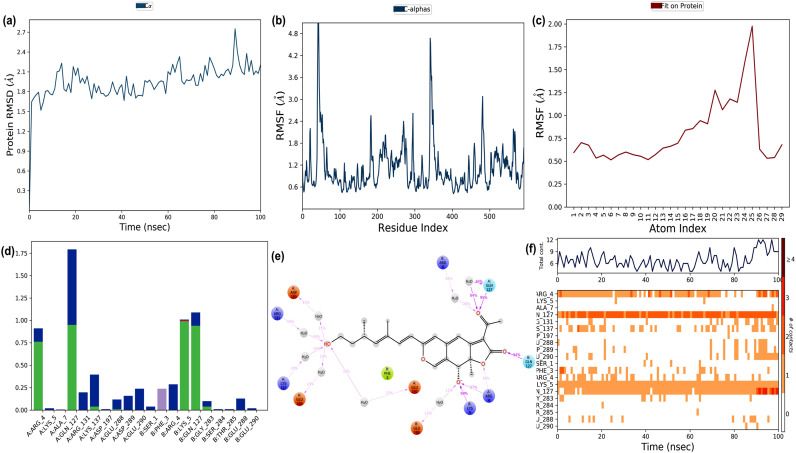
Fig. 10.
The binding and stability of the docked complex of natural lead molecule Rotiorinol-C and replicase polyprotein 1 ab of SARS-CoV-2 studied by molecular dynamic simulation at 100 ns? The trajectories obtained during the MD simulation is shown in figure (a) Protein-ligand RMSD: Protein RMSD (Å) is shown on the y-axis, and simulation time is given on the x-axis (b) Plot of the protein RMSF: RMSF (Å) is given in the y-axis and atom index is showed in the x-axis (c) Plot of ligand RMSF: RMSF (Å) is shown in y-axis and atom index is given in the x-axis (d) The protein-ligand contact is shown in the form of a histogram. The blue, grey, pink, and green regions in the histogram represent water bridges, hydrophobic interactions, ionic bonds, and hydrogen bonds respectively (e) The interactions between Hyoscyamine and post-fusion conformation of the spike glycoprotein observed during MD simulation (f) The protein-ligand contacts shown in the form of timeline representation. The top panel shows the total number of specific contacts between receptor and ligand, and the bottom panel shows the amino acid residues of the target, which is interacted with the ligand.