Table 3.
The binding potential of selected natural lead molecules towards the probable drug targets of human SARS-CoV-2 predicted by molecular docking studies.
| Protein name | PDB ID | Ligand names | Ligand structure | Binding affinity (Kcal/mol) | RMSD (Å) | Interacting residues | Hydrogen bonds |
|---|---|---|---|---|---|---|---|
| Spike glycoprotein | 1WYY | Hyoscyamine Pubchem ID: 154417 Source: Datura stramonium |
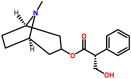 |
−8.14 | 0.0 | Ser924, Thr925, Gly928, Asp932, Gln1161, ILe1164, Asn1168 (Chain A), Ser924, Thr925, Gly928, Gln1161, Ile1164 (Chain B) | Asn1168: 2 |
| Spike glycoprotein-Closed state | 6VXX | −5.7 | 0.0 | Leu335, Pro337, Phe338, Gly339, Asp364, Val367 | 0 | ||
| Spike glycoprotein-Open state | 6VYB | −6.0 | 0.0 | Trp104, Ile119, Val126, Ile128, Tyr170, Ser172, Ile203, Val227 | Ser172: 1 | ||
| Membrane protein | 3I6G | −0.7 | 0.0 | Gly0 | 0 | ||
| Envelope small membrane protein | 2MM4 | −2.2 | 0.0 | Lys63, Asn64 | 0 | ||
| Replicase polyprotein 1 ab | 1Q2W | −6.1 | 0.0 | Glu156, Met165, Asp187, Arg188, Gln189, Gln192 | Gln192 | ||
| Replicase polyprotein 1a | 5RE4 | −6.0 | 0.0 | Trp218, Asn221, Leu271 | Trp218: 1 | ||
| Protein 7a | 1XAK | −4.4 | 0.0 | Tyr3, Tyr5, His47, Gln61 | 0 | ||
| Protein 9b | 2CME | −4.4 | 0.0 | Arg68, Ala69, Phe70 | 0 | ||
| Receptor binding domain of membrane protein | 6M17 | −5.4 | 0.0 | Thr376, Val407, Ala411, Val433, Tyr508 | 0 | ||
| Non-structural protein 7 | 6M71_C | −5.3 | 0.0 | Thr46, Phe49, Val53 | 0 | ||
| Non-structural protein 8 | 6M71_B, D | −5.9 | 0.0 | Leu128, Val130, Thr141, Tyr149 | 0 | ||
| Non-structural protein 12 | 6M71_A | −5.4 | 0.0 | Pro412, Phe415, Phe440, Phe843, | 0 | ||
| Non-structural Protein 6 | Hypothetical model | −5.2 | 0.0 | Asp6, Phe7, Leu15, Leu40, Ile60 | 0 | ||
| Non-structural Protein 10 | Hypothetical model | −6.1 | 0.0 | Ile55, Cys74, Tyr76, His83, Asp91, Leu112, Thr115, Val116 | His83:1 | ||
| Protein 3a | Hypothetical model | −5.3 | 0.0 | Phe56, Ile63, Lys66, Tyr189, Glu191, | 0 | ||
| Spike glycoprotein | 1WYY | Rotiorinol C Pubchem ID: 11703984 Source: Chaetorium cupreum |
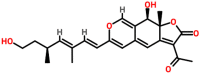 |
−9.82 | 0.0 | Arg4, Lys5, Tyr126, Gln127, Arg131, Asp289 and Glu290 | 5, Lys5, Gln127, Asp 289, Glu290 (A chain) Glu127 (B chain) |
| Spike glycoprotein-Closed state | 6VXX | −6.2 | 0.0 | Arg34, Thr208, Pro209, Leu212, Pro217, Gln218, Phe220 | Phe220: 1 | ||
| Spike glycoprotein-Open state | 6VYB | −6.3 | 0.0 | Thr33, Phe59, Asp287 | 0 | ||
| Membrane protein | 3I6G | −0.6 | 0.0 | Gly0 | 0 | ||
| Envelope small membrane protein | 2MM4 | −2.2 | 0.0 | Lys63, Asn64 | 0 | ||
| Replicase polyprotein 1 ab | 1Q2W | −7.0 | 0.0 | Phe3, Lys5, Arg131, Trp207, Phe291, Ile286, Asp289 | Trp207: 1 | ||
| Replicase polyprotein 1a | 5RE4 | −6.7 | 0.0 | Lys137, Thr199, Tyr239, Leu286, Leu287, Glu288, Asp289 | Tyr239, Asp289: 3 | ||
| Protein 7a | 1XAK | −5.4 | 0.0 | Tyr3, His47, Arg57, Thr59 | 0 | ||
| Protein 9b | 2CME | −5.0 | 0.0 | Ser56, Leu65, Glu66, Ala67, Arg68, Ala69, Phe70, Ser72 | Ser56: 1 | ||
| Receptor binding domain of membrane protein | 6M17 | −6.1 | 0.0 | Cys336, Phe338, Gly339, Ala363, Asp364, Val367, Ser371, Ser373, Phe374, | Ser373: 1 | ||
| Non-structural protein 7 | 6M71_C | −5.9 | 0.0 | Lys2, Asp5, Thr9, Thr46, Phe49, Val53, Met52 | 0 | ||
| Non-structural protein 8 | 6M71_B, D | −6.0 | 0.0 | Pro133, Asp134, Tyr135, Trp182, Pro178 | Tyr135: 1 | ||
| Non-structural protein 12 | 6M71_A | −6.6 | 0.0 | Arg181, Gln184, Asn213 | 0 | ||
| Non-structural protein 6 | Hypothetical model | −5.6 | 0.0 | Gln8, Lys38, Lys42, Leu44 | Lys42: 1 | ||
| Non-structural Protein 10 | Hypothetical model | −6.8 | 0.0 | Ile55, His83, Lys96, Val116 | His83:1 | ||
| Protein 3a | Hypothetical model | −6.3 | 0.0 | Phe28, Val29, Arg68, Ala72, Val90 | 0 | ||
| Spike glycoprotein | 1wyy | Scutifoliamide A Chemspider ID: 23311285 Source: Piper scutifolium |
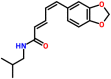 |
−6.4 | 0.0 | Asn910, Ala940, Thr943, Leu944, Gln947, Asn951, Leu1178, Leu1181, Glu1183 | 0 |
| Spike glycoprotein-Closed state | 6VXX | −5.4 | 0.0 | Leu118, Val120, Phe135, Leu141, Leu241 | 0 | ||
| Spike glycoprotein-Open state | 6VYB | −6.6 | 0.0 | Phe823, Asn824, Val826, Pro863, Pro1057, His1058 | Asn824: 1 | ||
| Membrane protein | 3I6G | −0.7 | 0.0 | Gly0 | 0 | ||
| Envelope small membrane protein | 2MM4 | −0.8 | 0.0 | Ly63 | 0 | ||
| Replicase polyprotein 1 ab | 1Q2W | −6.9 | 0.0 | Arg4, Lys5 and Phe291 (A chain), Lys 5 (B chain) | 1; Lys5 (B chain) | ||
| Replicase polyprotein 1a | 5RE4 | −6.8 | 0.0 | Val212, Arg217, Leu220, Gln256, Ile259, Asp263 | 0 | ||
| Protein 7a | 1XAK | −5.6 | 0.0 | Gln6, Cys8, Val9, Thr12, Leu16, Lys17 | Val9, Lys17: 2 | ||
| Protein 9b | 2CME | −4.4 | 0.0 | Ala58, Arg68, Ala69, Phe70, Ser72 | 0 | ||
| Receptor binding domain of membrane protein | 6M17 | −5.9 | 0.0 | Gln493, Tyr495, Gly502, Tyr505 | Gln493, Gly502: 2 | ||
| Non-structural protein 7 | 6M71_C | −5.9 | 0.0 | Asp5, Thr9, Thr46, Phe49, Glu50, Val53 | Thr9: 1 | ||
| Non-structural protein 8 | 6M71_B, D | −6.0 | 0.0 | Leu128, Met129, Pro133, Thr141, Tyr149 | 0 | ||
| Non-structural protein 12 | 6M71_A | −6.1 | 0.0 | Asp484, Ile488, Gln573, Ser578, Ala581 | 0 | ||
| Non-structural Protein 6 | Hypothetical model | −5.5 | 0.0 | Leu16, Ile17, Arg20, Thr21 | Arg20: 1 | ||
| Non-structural Protein 10 | Hypothetical model | −5.6 | 0.0 | His83, Leu92, Asn114, Thr115, Val116 | Val116: 2 | ||
| Protein 3a | Hypothetical model | −5.4 | 0.0 | Cys130, Trp131, His150, His204, His227 | 0 | ||
| Spike glycoprotein | 1wyy | Tamaridone Pubchem ID: 15345466 Source: Tamarix dioica |
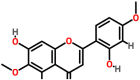 |
−7.2 | 0.0 | Asp931, Asn935, Lys1162, Asp1165, Asn1168, Lys1172 | 1; Asn935 |
| Spike glycoprotein-Closed state | 6VXX | −7.3 | 0.0 | Ala520, Phe559, Phe562, Gln563, Gln564, Phe565, Gly566, Arg567 | Gln564, Phe565, Arg567: 4 | ||
| Spike glycoprotein-Open state | 6VYB | −6.8 | 0.0 | Thr732, Thr778, Ser780, Pro863, His1058 | Ser730: 2 | ||
| Membrane protein | 3I6G | −0.8 | 0.0 | Gly0 | 0 | ||
| Envelope small membrane protein | 2MM4 | −3.1 | 0.0 | Lys63, Asn64 | Asn64: 1 | ||
| Replicase polyprotein 1 ab | 1Q2W | −6.3 | 0.0 | Gly11, Lys12, Glu14, Gly15, | Gly11: 1 | ||
| Replicase polyprotein 1a | 5RE4 | −7.0 | 0.0 | Leu87, Lys137, Thr190, Tyr237, Tyr239, Leu285 | Tyr237: 1 | ||
| Protein 7a | 1XAK | −5.7 | 0.0 | Val9, Thr12, Val14, Arg65 | 0 | ||
| Protein 9b | 2CME | −5.6 | 0.0 | Arg68, Gln71, Ser72 | 0 | ||
| Receptor binding domain of membrane protein | 6M17 | −6.5 | 0.0 | Thr345, Arg346, Ser349, Lys441, Lys444, Asn448, Asn450, Tyr451 | Lys444: 1 | ||
| Non-structural protein 7 | 6M71_C | −5.9 | 0.0 | Lys2, Val6, Thr9, Phe49, Met52 | Thr9: 1 | ||
| Non-structural protein 8 | 6M71_B, D | −5.7 | 0.0 | Leu128, Val130, Val131, Thr141, Tyr149 | 0 | ||
| Non-structural protein 12 | 6M71_A | −6.7 | 0.0 | Lys47, His133, Asn138, Lys780 | 0 | ||
| Non-structural Protein 6 | Hypothetical model | −5.6 | 0.0 | Lys48, Ser50, Leu52, Asp53, Gln56 | Leu52: 1 | ||
| Non-structural Protein 10 | Hypothetical model | −6.7 | 0.0 | Pro37, Asn105, Asp106, Phe110, Lys113, Asn114, Tyr126 | Asn105, Asn114: 2 | ||
| Protein 3a | Hypothetical model | −6.6 | 0.0 | Gln213, Val255, Ile263, Thr270 | 0 |
