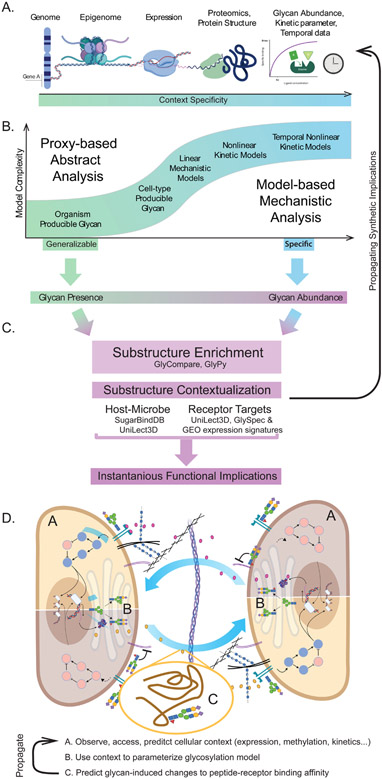
Figure 5 -. The progression of model complexity and predictions appropriate, given various common data types.
(A) Various datatypes of increasing complexity and rarity necessary to train different models. (B) The relationship between model specificity and model complexity. As the complexity increases and the magnitude and rarity of input data increases, so does the specificity of the model. Though lower complexity models are not as specific, they can be beneficially generalizable. (C) Finally, once the glycoprofile predictions are complete, they can be compared to lectin and substructure databases to predict what receptor-ligand interactions the differential glycosylation may impact. Panel D describes two theoretical cells (Figure 3C) co-regulating through differential glycosylation. If a reasonable differential expression signature can be inferred (due to differential interference or promotion of a receptor-ligand interaction), we can generate a “gene-availability” based prediction of updated glycosylation, thus exploring the sustainability of a glycosylation pattern. Labels A, B, and C correspond to methods illustrated in panels in A, B, and C respectively: (A) The omics assessment of the intracellular environment, (B) modeling of the glycan biosynthetic implications of the intracellular environment, (C) differential glycosylation of peptides and receptors and (A-C) the propagation of that information to the intercellular interface.