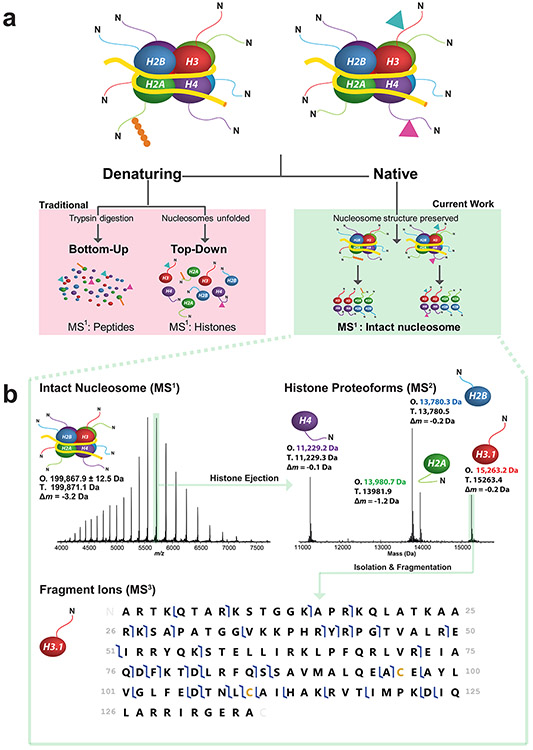
Figure 1. Three strategies of histone analysis, including Nuc-MS for the direct interrogation of intact nucleosomes.
(a) In the figure’s hypothetical nucleosome mixture (a, top), the Nuc-MS workflow (a, right side) detects the co-localization of H3 and H4 methylation (blue and pink triangles) and determines that the H2A ubiquitination (in orange) is present on a separate nucleosome. In contrast, traditional methods (a, left side) use either protease-derived peptides or whole histones under denaturing conditions to detect histone PTMs, blurring the modification states of intact nucleosomes in a mixture. Nuc-MS detects proteoforms and their PTMs present in intact nucleosomes by employing top-down MS in native mode (a, right side). (b) Data from the three steps of Nuc-MS on an intact unmodified nucleosome. First, the mass of intact nucleosomes is measured (MS1: O, observed average mass; T, theoretical mass; Δm, error). Second, a single nucleosome charge state (e.g. 35+ ions highlighted in green) is isolated and activated by collisions with nitrogen to eject all intact histones and detect them simultaneously at isotopic resolution (MS2, reporting monoisotopic masses). Third, each histone is isolated and further activated to create backbone fragmentation products that characterize the proteoforms, revealing PTMs or sequence events (MS3; blue flags indicate fragment ions matching uniquely to human histone H3.1, depicted as a graphical fragment map at bottom; green rectangle in the upper right of the panel highlights the intact precursor).