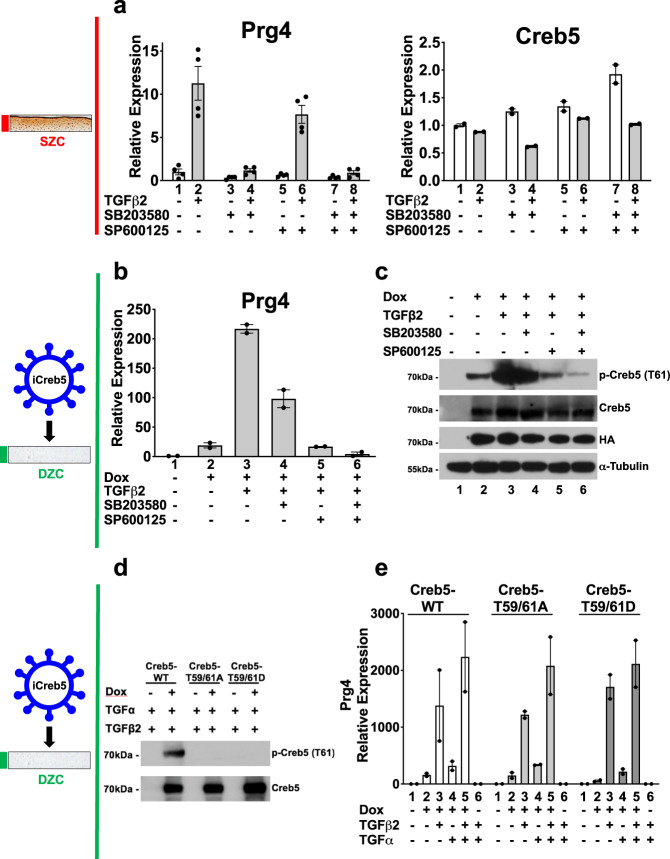
Fig. 5. Creb5-dependent induction of Prg4 requires SAPK activity, but neither phosphorylation of T59 nor T61 in Creb5.
a TGF-β2 induced expression of Prg4 in superficial zone articular chondrocytes is blocked by inhibition of p38 activity. SZCs were cultured in either the absence (white) or presence (gray) of TGF-β2 (20 ng/ml), plus a p38 inhibitor (SB203580; 10 μM), or a JNK inhibitor (SP600125; 10 μM), as indicated for 2 days. Gene expression was assayed by RT-qPCR and normalized to Gapdh. Error bar indicates standard error of the mean (n = 4 technical repeats for Prg4 expression, and n = 2 technical repeats for Creb5 expression). Similar results have been obtained in three independent biological repeats. b, c DZCs were infected with a lentivirus encoding doxycycline-inducible Creb5-HA. After selection in G418, the cells (iCreb5-DZCs) were cultured in either the absence or presence of doxycycline (1 μg/ml), TGF-β2 (20 ng/ml), a p38 inhibitor (SB203580; 10 μM), or a JNK inhibitor (SP600125; 10 μM), as indicated for 2 days. b Gene expression was assayed by RT-qPCR and normalized to Gapdh. Error bar indicates standard error of the mean (n = 2 technical repeats). Similar results have been obtained in three independent biological repeats. c Western analysis of exogenous Creb5-HA expression and phosphorylation in iCreb5-DZCs with antibodies directed against either phospho-Creb5 (T61), total Creb5, HA, or α-tubulin. Similar results have been obtained in two independent biological repeats. d, e DZCs were infected with a lentivirus encoding either iCreb5-WT-HA; iCreb5-T59,61A-HA; or iCreb5-T59,61D-HA. After selection in G418, the cells were cultured in either the absence or presence of doxycycline (1 μg/ml), TGF-β2 (20 ng/ml), and TGF-α (100 ng/ml) as indicated for 3 days. d Western analysis of iCreb5-WT/mutant expression in iCreb5-DZCs with antibodies directed against either phospho-Creb5 (T61) or total Creb5. Similar results have been obtained in two independent biological repeats. e Gene expression was assayed by RT-qPCR and normalized to Gapdh. Error bar indicates standard error of the mean (n = 2 technical repeats). Similar results have been obtained in three independent biological repeats.