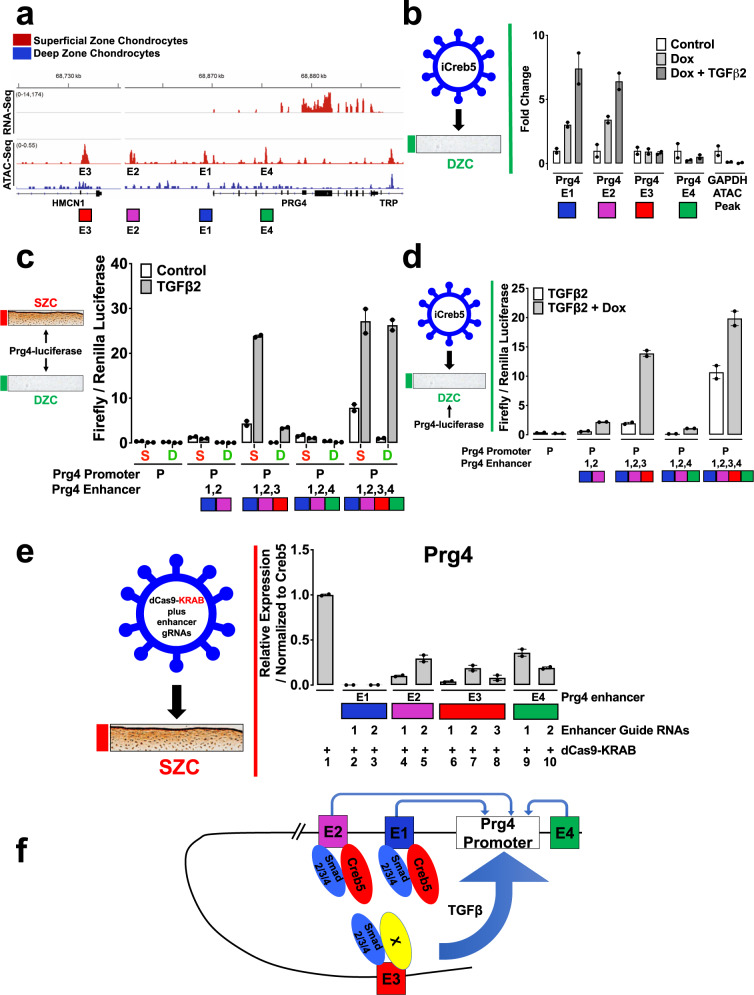
Fig. 7. Prg4 expression is regulated by the combinatorial activity of several regulatory elements.
a Comparison of RNA-Seq (top tracks) and ATAC-Seq (bottom tracks) surrounding the Prg4 locus. Signals for either SZCs (red) or DZCs (blue) are displayed. Putative Prg4 regulatory elements (E1–E4) are indicated. b Bovine articular chondrocytes were infected with lentivirus encoding doxycycline-inducible HA-tagged Creb5 (iCreb5-HA). The cells were cultured in either the absence or presence of doxycycline and TGF-β2, as indicated. ChIP-qPCR was performed with anti-HA. Creb5-HA occupancy on ATAC-Seq peaks E1–E4 in the Prg4 locus or in the Gapdh locus are displayed. Error bar indicates standard error of the mean (n = 2 technical repeats). Similar results have been obtained in two independent biological repeats. c SZC-enriched ATAC-Seq peaks surrounding the Prg4 locus work in combination. Either superficial zone (S) or deep zone (D) bovine articular chondrocytes were co-transfected with a firefly luciferase reporter driven by both the Prg4 promoter plus a combination of enhancers (E1–E4) surrounding this gene and a CMV-renilla luciferase construct. The cells were cultured in either the absence or presence of TGF-β2, as indicated. Relative expression of firefly/renilla luciferase is displayed. Error bar indicates standard error of the mean (n = 2 technical repeats). Similar results have been obtained in three independent biological repeats. d DZCs were infected with a lentivirus encoding doxycycline-inducible Creb5-HA. After selection in G418, the cells (iCreb5-DZCs) were transfected with Prg4-firefly luciferase expression constructs as described above and cultured with TGF-β2 (20 ng/ml) in either the absence or presence of doxycycline (1 μg/ml), as indicated for 2 days. Relative expression of firefly/renilla luciferase is displayed. Error bar indicates standard error of the mean (n = 2 technical repeats). Similar results have been obtained in two independent biological repeats. e SZCs were infected with lentivirus encoding only dCas9-KRAB or with lentivirus programmed to encode both dCas9-KRAB plus guide RNAs targeting the various SZC-enriched ATAC-seq peaks (two to three different guides for each ATAC-Seq peak) that surround the Prg4 locus. After selection in puromycin, the cells were cultured in the presence of TGF-β2 (20 ng/ml) for 3 days. Gene expression was assayed by RT-qPCR and normalized to Creb5. Relative levels of Prg4 expression in cells expressing dCas9-KRAB plus various guide RNAs (lanes 2–10) are compared to cells expressing only dCas9-KRAB (lane 1). Error bar indicates standard error of the mean (n = 2 technical repeats). Similar results have been obtained in three independent biological repeats. f Model for Creb5-dependent induction of Prg4 expression.