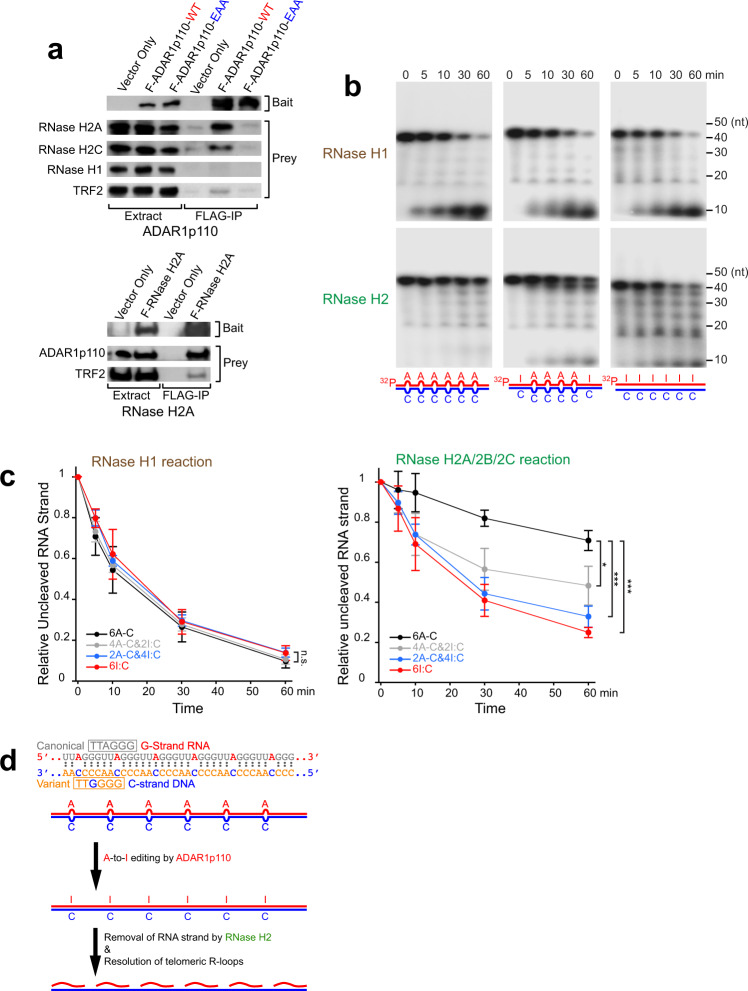
Fig. 8. Editing of A–C mismatches in telomeric repeat RNA:DNA hybrids facilitates RNase H2 cleavage of RNA strands.
a F-ADAR1p110-WT recombinant protein was copurified with endogenous RNase H2 subunits H2A and H2C, but not with RNase H1. No RNase H2 association detected with a dsRNA-binding defective mutant F-ADAR1p110-EAA (upper panels). Similarly, FLAG-RNase H2A pulled down endogenous ADAR1p110 (lower panels). Note that both FLAG-AFAR1p110 and FLAG-RNase H2A pulled down endogenous TRF2, a telomere-binding protein and a member of the Shelterin complex, supporting our hypothesis that ADAR1 and RNase H2 collaborate together to resolve telomeric R-loops. Protein molecular weight markers are presented in Source Data file. b In vitro assay for digestion of 5′-32P-labeled RNA strands of telomeric repeat RNA:DNA hybrids by RNase H1 or RNase H2A/2B/2C complexes. RNase H2 could not degrade the RNA strands of telomeric repeat RNA:DNA hybrids containing six A–C mismatches, but began digesting RNA strands of RNA:DNA hybrids containing four A–C mismatches. Replacement of all A–C mismatches with I:C-matched base pairs resulted in efficient digestion of RNA strands by RNase H2. RNase H1 degraded RNA strands regardless of the number of A–C mismatches. c Time course of digestion of RNA strands of telomeric repeat RNA:DNA hybrids with varying numbers of A–C mismatches by RNase H1 or RNase H2. Data are mean ± SD (n = 4, technical replicates); significant differences were identified by two-tailed Student’s t tests: *P < 0.05, ***P < 0.001, n.s., not significant. All individual experimental data values and exact P values are presented in Supplementary Data 3. d A-to-I editing of A–C mismatches to I:C base pairs in RNA:DNA hybrids facilitates digestion of RNA strands by RNase H2.