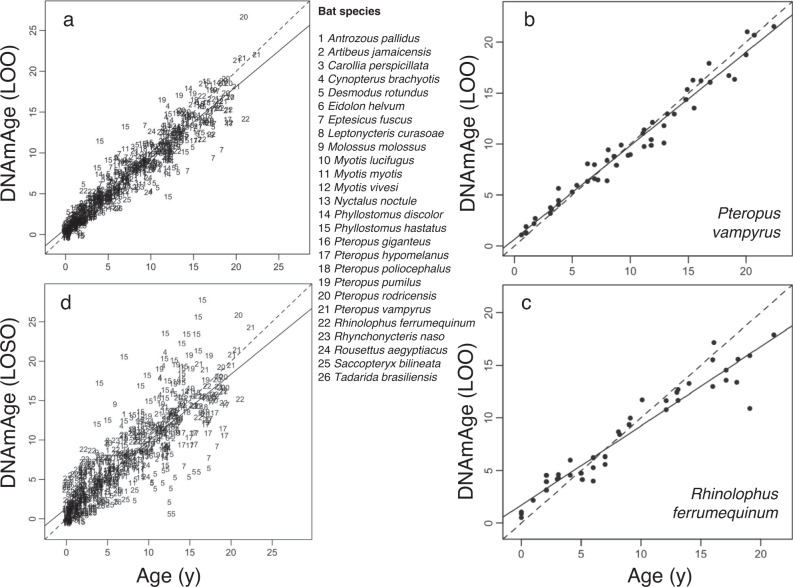
Fig. 1. Epigenetic clocks accurately predict the chronological age of bats.
a Leave-one-out (LOO) cross-validation based on penalized regression gave a correlation of 0.95 with a median absolute error (MAE) of 0.74 years between observed and predicted (DNAmAge) age (after square-root transform) for 26 bat species. To ensure an unbiased cross-validation analysis, we allowed the number of CpGs to change with the respective training data. b LOO cross-validation based on penalized regression of 51 Pteropus vampyrus samples gave a correlation of 0.99 with MAE of 0.72 years between observed and predicted age. c LOO cross-validation based on penalized regression of 40 Rhinolophus ferrumequinum samples gave a correlation of 0.96 with MAE of 1.11 years between observed and predicted age. d Cross-validation analysis in which the DNAm data for one species was left out (LOSO) and ages are predicted for that species using a clock estimated with the remaining data. The resulting correlation between observed and predicted age is 0.84 (MAE = 1.41 years). Additional epigenetic clocks for individual species and genera are in Supplementary Figs. 1 and 2.