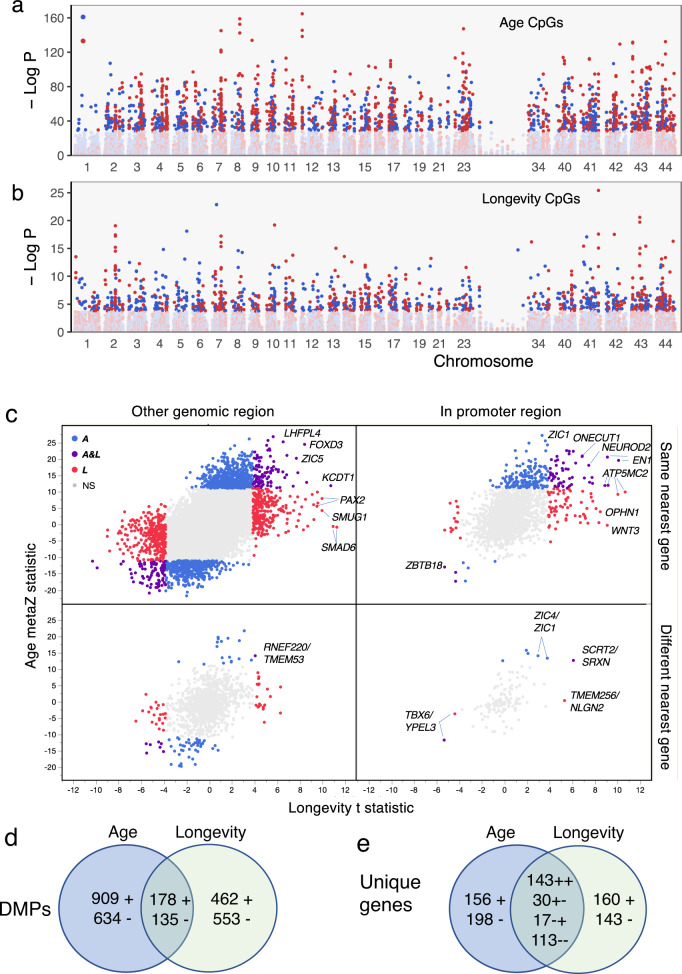
Fig. 3. Differentially methylated positions (DMPs) for age and longevity are widely distributed and partially overlap.
a Negative log P for age-associated DMPs plotted against location on each Rhinolophus ferrumequinum chromosome. The top 2000 age-associated DMPs are darkened with increasing DNAm indicated by red and decreasing DNAm indicated by blue. Hypermethylated age-DMPs are underrepresented on chromosome 1, which is syntenic with the human X chromosome. b Longevity DMPs are also distributed across all R. ferrumequinum chromosomes. Darkened symbols indicate 1491 significant (BY 5% FDR) longevity DMPs with colors indicating DNAm direction as in (a). c Effect of DNAm change on age plotted against the effect of DNAm change on longevity (see “Methods”) illustrates the association between age and longevity effects. Significant sites are colored blue for age, red for longevity, and purple for both age and longevity. Symbols for the orthologous gene with the nearest transcription start site (TSS) to the DMP are indicated for a sample of extreme age and longevity DMPs. Bottom panels indicate DMPs that map to different genes in the short-lived species, M. molossus, and the long-lived species, R. ferrumequinum, with the M. molossus gene indicated after /. Note that most extreme age and longevity DMPs in promoter regions (i.e., −10,000 to +1000 bp from the TSS) are in the upper right panel, i.e., nearest the same gene in both species. d Age DMPs overlap 17% with hypermethylating (+) and hypomethylating (−) longevity DMPs in M. molossus. Long-lived bat species show similar patterns (Supplementary Fig. 5a). e Number of unique genes nearest age and longevity DMPs for M. molossus. Signs on numbers in the overlap region indicate methylation direction for age then longevity. Long-lived bat species show similar patterns (Supplementary Fig. 5b).