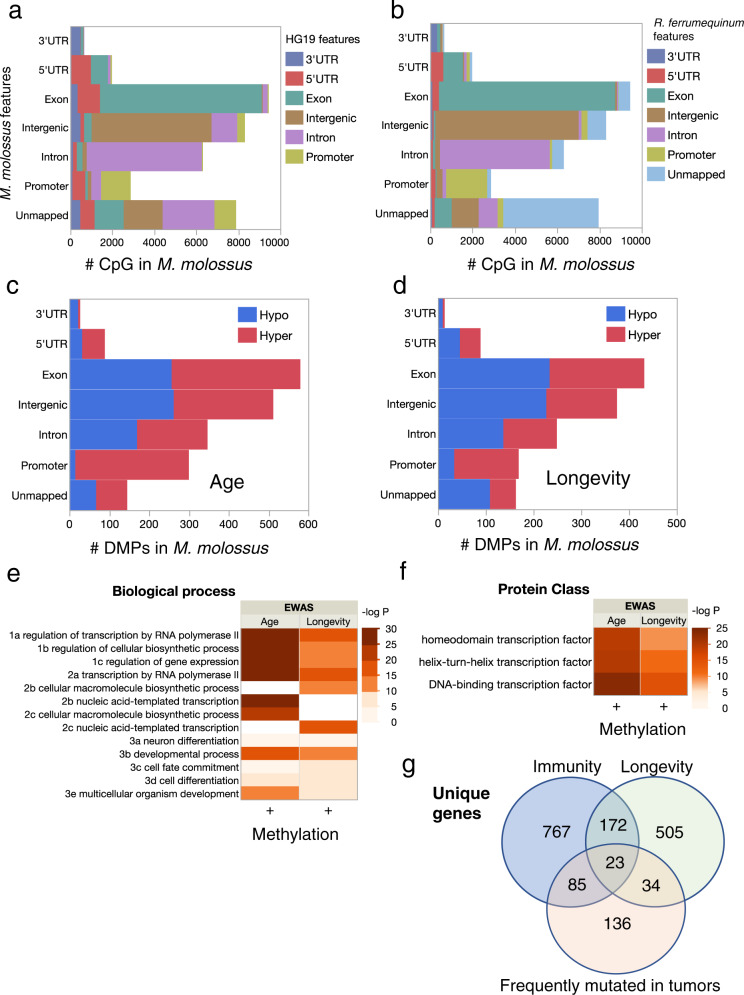
Fig. 4. Age and longevity DMPs are enriched in promoter regions of genes associated with immunity and cancer.
a CpG annotation for the short-lived bat, M. molossus, in comparison to genome regions where probes map to the human genome (HG19) shows that fewer than half of the probes that map to a promoter region in the bat also map to a promoter region in human (see also Supplementary Fig. 4). Colors indicate genomic regions in the human genome as indicated in the legend. b In contrast, CpG annotation comparison between two phylogenetically distant bat species, M. molossus, and R. ferrumequinum, indicates greater probe conservation with respect to gene proximity (see also Supplementary Fig. 4). Colors indicate genomic regions in the genome of R. ferrumequinum. c The top 2000 age DMPs are highly enriched near promoter regions with over 95% exhibiting hypermethylation in M. molossus and other bats (Supplementary Fig. 5). Red indicates DMPs associated with increasing DNAm, blue indicates DMPs associated with decreasing DNAm. d The 1491 longevity DMPs are also enriched in promoter regions with over 80% exhibiting hypermethylation in M. molossus and other bats (Supplementary Fig. 5). Color as in (d). e Enriched biological processes for unique M. molossus genes from promoter regions are only significant for hypermethylating age and longevity DMPs. Only three significant GO terms from each parent–child group are shown to minimize redundancy. f Enrichment analysis of protein class for unique M. molossus genes from promoter regions reveals significant enrichment of helix-turn-helix transcription factors (TF) only for hypermethylated DMPs associated with age and longevity. Cell color indicates significance (negative log P for GO terms with adjP < 10e−4) of enrichment in (e) and (f). g Overlap between genes associated with longevity, innate immunity, or frequently mutated in human tumors identified in M. molossus. Enrichment analyses using genome annotations from other bat species produce similar results (Supplementary Fig. 6).