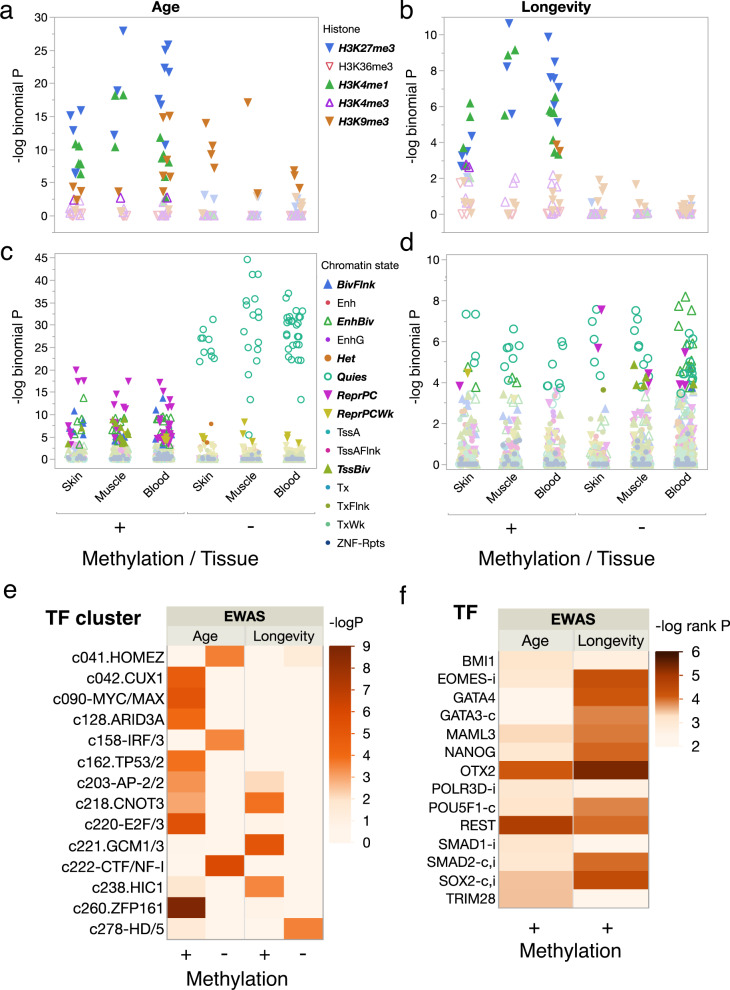
Fig. 5. Functional overlap analysis of DMPs reveals the role of key transcriptional regulators.
Histone marks (e.g., H3K27me3 = trimethylation of lysine 27 on histone H3) are denoted by shape and color as indicated in the legend for cell lines derived from skin, muscle, or blood for DMPs mapped in Desmodus rotundus, a long lifespan species, for (a) age and (b) longevity with darkened symbols indicating significance (BY 5% FDR) and ±indicating positive/negative rates of DNAm change. Enriched chromatin states (e.g., ReprPC = Repressed Polycomb) as predicted by a hidden Markov model for cell lines derived from skin, muscle, or blood are denoted by shape and color as indicated in the legend for (c) age and (d) longevity DMPs mapped in Desmodus rotundus with darkened symbols indicating significance (BY 5% FDR) and ±indicating positive/negative rates of DNAm change. e Transcription factor clusters enriched for hypermethylated (+) and hypomethylated (−) age or longevity DMPs with cell color indicating significance (negative log P, adjP < 10e−4) of overlap with predicted transcription factor binding sites in probe sequences using a hypergeometric test. f Top-ranked transcription factors associated with the change in expression of genes containing age or longevity DMPs in promoter regions in M. molossus, with integrative rank significance (see “Methods”) indicated as negative log P. Genes frequently mutated in human tumors are indicated by (c), and those involved in innate immunity by i. Only genes with hypermethylated sites in promoter regions showed evidence of enrichment. Analyses using genome annotations from other bat species produce similar results (Supplementary Fig. 6).