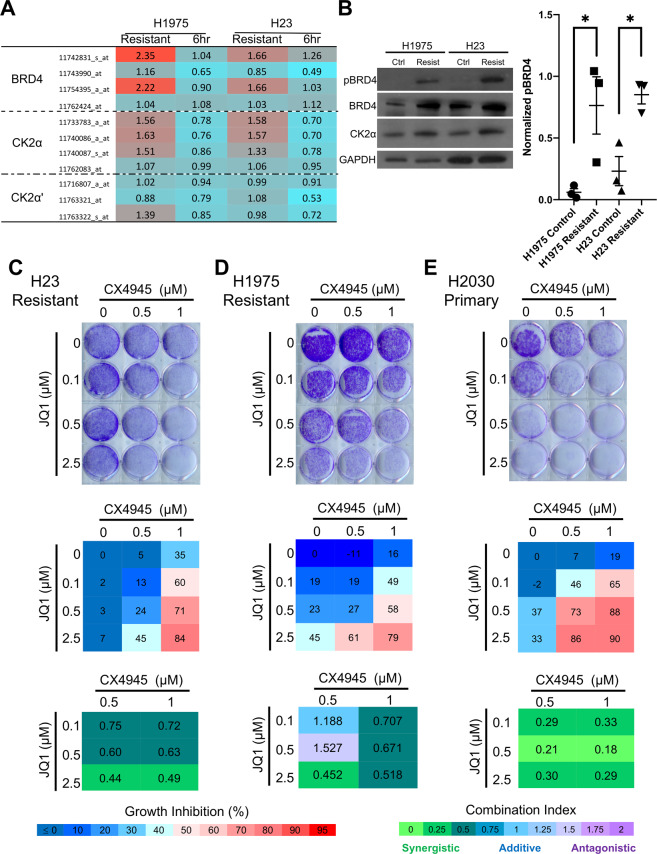
Fig. 5. Phosphorylated BRD4 and the kinase CK2 are elevated in JQ1 resistant lines and are synergistic in inducing cell death when inhibited.
A mRNA expression analysis shows increased expression of BRD4 and CK2α levels for resistant lines, but no change in CK2α′ levels. Values represent fold changes as compared to the control line for each specific cell line and condition. 11742831_s_at and 11754395_a_at are probes specific for the long isoform of BRD4. Color scale represents lower (blue) or higher (red) degree of expression compared to control. B Western blot analysis of pBRD4 and BRD4 levels, specific to the long isoform, as well as CK2α levels in JQ1 resistant and their control counterparts. Densitometry data for relative pBRD4 levels are plotted in scatter format for three independent western blots from independent cell lysates. GAPDH serves as a loading control and bars represent SEM (*P < 0.05, two-tailed, unpaired T test). C–E Combination growth assays for H23 (C) and H1975 (D) acquired resistant lines, and H2020 (E) primary resistant LAC cell line treated with the indicated combinations of JQ1 and CX-4945 for 10 days. Crystal violet images are representative of triplicate biological experiments. Cell viability was measured at the endpoint using Alamar Blue and the percent inhibition calculated compared to the no drug control, with the mean value of three biological replicates indicated. CompuSyn was used to calculate combination index (CI) scores for each drug combination with <0.75 indicating synergy, 0.75–1.25 indicating additive effects, and >1.25 indicating antagonism. Legends for percent growth inhibition and combination index values are shown at the bottom.