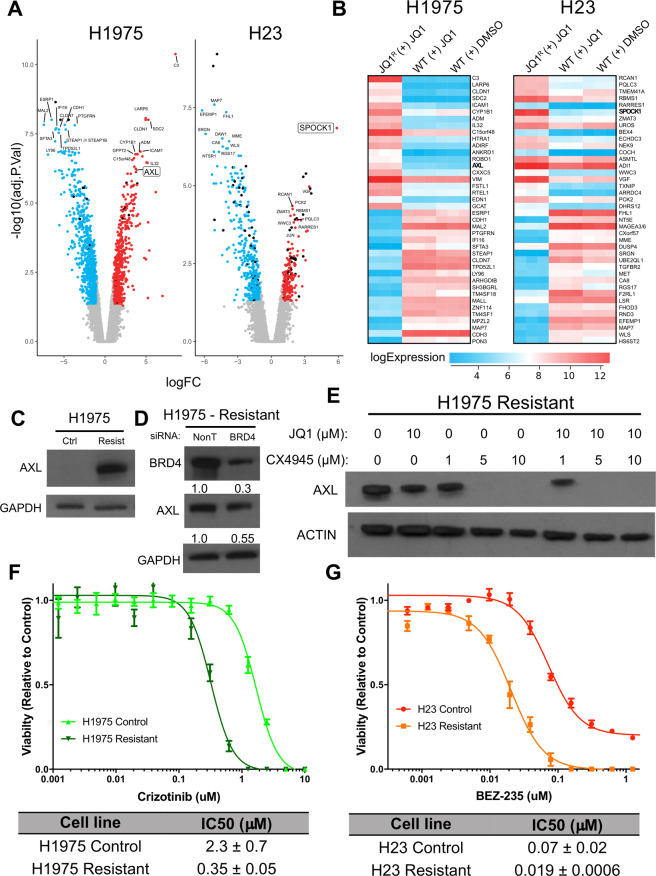
Fig. 6. AXL and SPOCK1 are highly upregulated in JQ1 resistant lines and potential downstream effector targets of pBRD4.
A Volcano plots for genes differentially expressed specifically in JQ1 resistant H23 (left) and H1975 (right) cells. Expression profiles were generated in triplicate as described in methods. Resistant cells and control cells treated with 10 μM JQ1 for 6 h were compared to control cells to identify significantly differentially expressed genes (>2-fold change, BH corrected p < 0.05, LIMMA) and the resulting lists compared to identify those deregulated only in the resistant lines. Genes significantly underexpressed (blue) or overexpressed (red) are indicated with selected gene names indicated. SPOCK1 and AXL are highlighted with a box around the gene name in H23 and H1975, respectively. B The top 20 up- and downregulated genes specific to the H1975 (left) and H23 (right) resistant lines. Expression values are plotted as a heatmap for the resistant line (JQ1R + JQ1), control cells treated with 10 μM JQ1 for 6 h (WT + JQ1) and control cells in vehicle only (WT + DMSO). Values from triplicate experiments are shown for each condition. The color indicates higher (red) or lower (blue) expression (see legend bar). C Validation of AXL overexpression in H1975 resistant cells by immunoblot. Note, GAPDH is the same as in Fig. 3B. D siRNA mediated knockdown of BRD4 reduces expression of AXL in H1975 resistant cells. Protein levels are normalized to the NonT siRNA control condition. The blots for BRD4 and GAPDH are the same as in Fig. 4D. E Inhibition of CK2 with CX-4945 decreases the expression of AXL in H1975 resistant cells. Cells were treated with the indicated doses of CX-4945 and JQ1 for 6 h before lysis and blotting for the indicated proteins. Actin serves as a loading control. F Crizotinib dose response assays (72 h) in H1975 resistant and control cells. G BEZ-235 dose response assays (72 h) in H23 resistant and control cells. For (F, G), graphs are representative of a single experiment while IC50 values represent means from two biological replicates (±SEM).