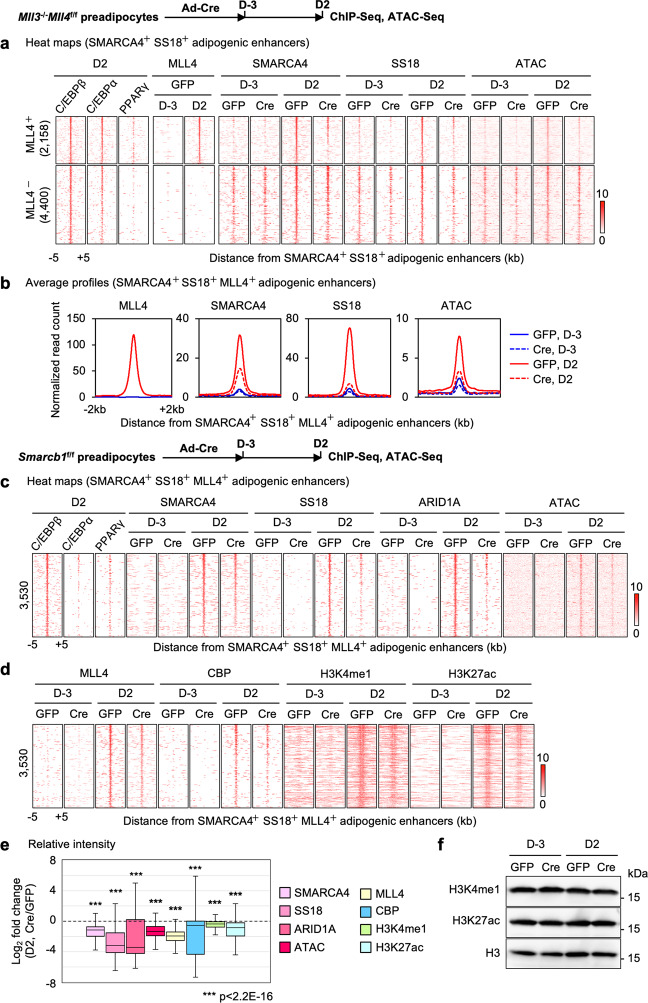
Fig. 5. Reciprocal regulation between BAF and MLL4 on active enhancers during adipogenesis.
Mll3‒/‒Mll4f/f or Smarcb1f/f brown preadipocytes were infected with adenoviruses expressing GFP or Cre, followed by adipogenesis assays. Cells were collected before (D-3) and during (D2) adipogenesis for ChIP-Seq, ATAC-Seq and Western blot analyses. a, b Decreased SMARCA4 and SS18 binding and chromatin accessibility on adipogenic enhancers in Mll4 KO (Cre) cells during adipogenesis. Adipogenic enhancers were defined as active enhancers bound by either C/EBPβ, C/EBPα, or PPARγ at D2 of adipogenesis. Published ChIP-Seq data sets for C/EBPβ, C/EBPα, and PPARγ were used (GSE74189)18. Heat maps around SMARCA4+ SS18+ adipogenic enhancers (a) and average profiles around SMARCA4+ SS18+ MLL4+ adipogenic enhancers (b) are shown. Adipogenic enhancers shown in the heat maps were ranked by the binding intensity of SMARCA4 at the center in D2 control (GFP) cells. c–f Deletion of Smarcb1 reduces binding of BAF subunits (SMARCA4, SS18, and ARID1A) as well as chromatin opening, and impairs MLL4 binding and activation of adipogenic enhancers. Adipogenic enhancers at D2 of adipogenesis were re-defined using ChIP-Seq data sets obtained in Smarcb1f/f cells. c Heat maps for ChIP-Seq of BAF subunits (SMARCA4, SS18, and ARID1A) and ATAC-Seq around the center of SMARCA4+ SS18+ MLL4+ adipogenic enhancers. d Heat maps for ChIP-Seq of MLL4, CBP, H3K4me1 and H3K27ac on adipogenic enhancers. Adipogenic enhancers shown in the heat maps were ranked by the intensity of SMARCA4 at the center in D2 control (GFP) cells. e Fold changes of intensities between control (GFP) and Smarcb1 KO (Cre) cells on adipogenic enhancers (n = 3530) are shown in box plots. The box represents the first and third quartiles with the horizontal line showing the median, and whiskers indicating minimum and maximum in all box plots. Outliers were not included. Statistical significance levels are as follows (Wilcoxon signed rank test, one-sided): SMARCA4 (p = 0); SS18 (p = 0); ARID1A (p = 6.3E-101); ATAC (p = 0); MLL4 (p = 4.5E-281); CBP (p = 0); H3K4me1 (p = 0); H3K27ac (p = 0). Exact p-values cannot be computed for SMARCA4, SS18, ATAC, CBP, H3K4me1 and H3K27ac due to ties in the ranks. f Western blot analyses of H3K4me1 and H3K27ac in control and Smarcb1 KO cells during adipogenesis. Total histone H3 was used as a loading control. n = 3 biological replicates.