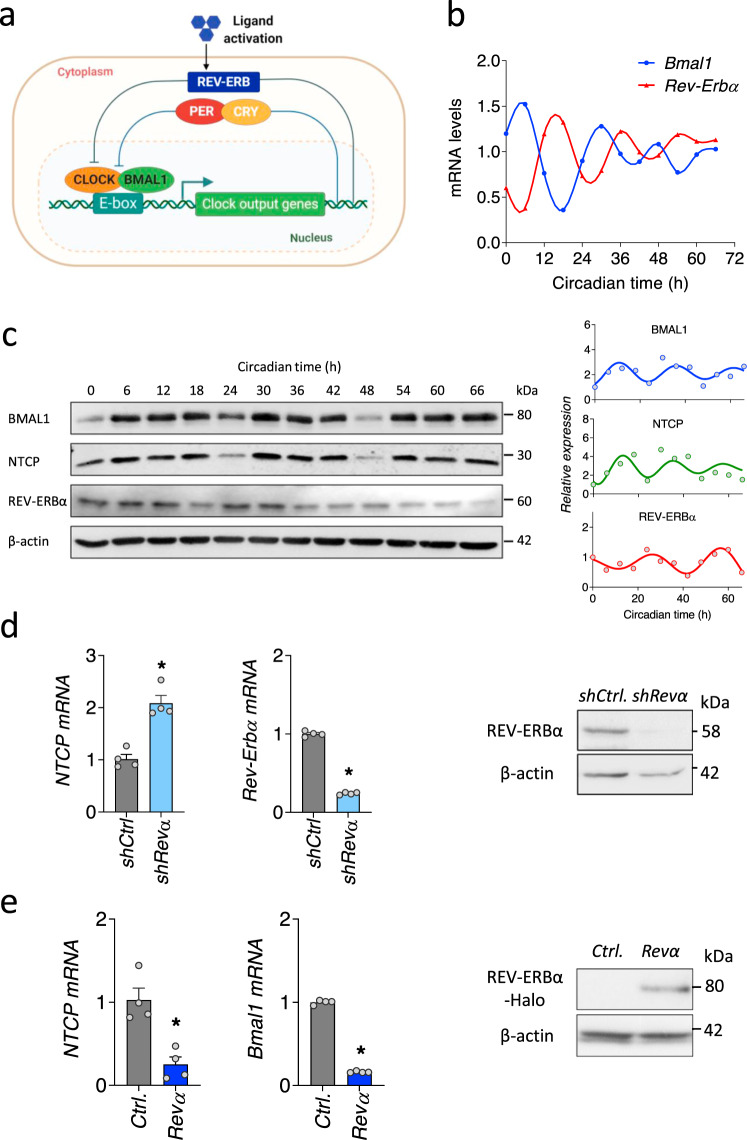
Fig. 1. REV-ERBα coordinates the circadian expression of NTCP.
a Circadian transcription–translation feedback loops generate daily rhythmic expression of circadian genes. The molecular clock generates a cycle of 24-hour periodicity when a heterodimeric transcription factor BMAL1-CLOCK activates the transcription from E-box sequences in target gene promoters. Gene products feedback to repress the transcriptional activity of the heterodimeric activator. Ligand activation of REV-ERB represses the transcription of Bmal1. b Synchronization of differentiated HepaRG cells (dHepaRG). dHepaRG cells were treated with dexamethasone at 100 nM for 2 h and samples collected at 6 h intervals. Bmal1 and Rev-Erbα mRNAs were measured by quantitative reverse transcription polymerase chain reaction (qRT-PCR) and expressed relative to the mean. Data are the average of two independent experiments. c Synchronized dHepaRG cells were assessed for BMAL1, NTCP and REV-ERBα expression together with housekeeping β-actin by western blotting. Densitometric analysis quantified BMAL1, NTCP and REV-ERBα in individual samples and was normalized to their own β-actin loading controls. Data are the average of two independent experiments. d Total RNA was extracted from control or Rev-Erbα silenced HepaRG cells and NTCP and Rev-Erbα mRNA levels measured by qRT-PCR. REV-ERBα expression together with housekeeping gene β-actin were assessed by western blotting. Data are expressed relative to control (mean ± SEM, n = 4, Mann–Whitney test, Two-sided). e Total RNA was extracted from control or Halo-tagged Rev-Erbα overexpressed HepaRG cells and Bmal1 and NTCP mRNA levels measured by qRT-PCR. Data are expressed relative to control (mean ± SEM, n = 4, Mann–Whitney test, Two-sided). *p < 0.05. Data are provided in the accompanying Source Data file.