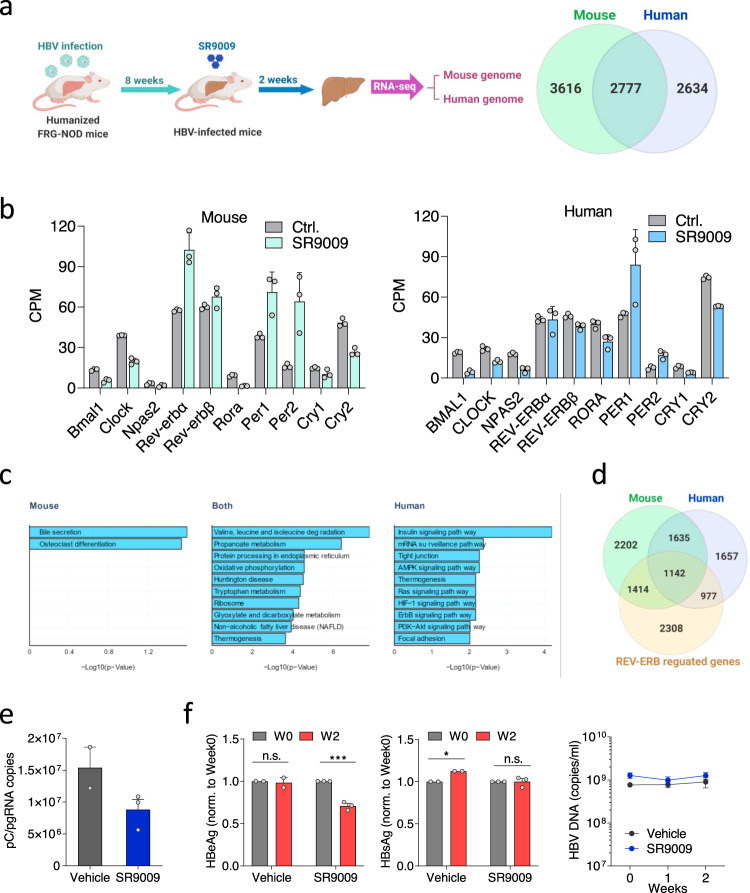
Fig. 6. Pharmacological activation of REV-ERB modulates the human transcriptome and inhibits HBV BCP transcription in vivo.
a Fah−/−, Rag2−/−, IL2Rg−/−, non-obese diabetic (FRG-NOD) mice transplanted with primary human hepatocytes were infected with 109 HBV genome equivalents purified from concentrated supernatant of the HepG2.2.15 cell line (HBV genotype D subtype ayw). Eight weeks later mice were treated twice a day by intraperitoneal injection of either vehicle or 100 mg/kg SR9009 for 2 weeks. At the end of experiment, mouse livers were collected for RNA-seq analysis and sequence reads mapped to the human or mouse genome. Differential expressed genes (DE) in human and mouse cells (19 of the DE genes in mouse had no human ortholog match). b Differentially expressed core clock genes in mouse and human. The counts per million (CPM) of the key clock genes assigned to the two genomes in the control and treated group are shown (mean ± SD, n = 3 independent liver samples). c Human KEGG pathway analysis of differentially expressed genes in mouse only, both mouse and human, and human only sets. Negative log10 adjusted p-values are given and top 10 significant (adjusted p-value <0.05) pathways shown. d Overlap of differentially expressed genes from SR9009 treated mouse livers with REV-ERB regulated genes. e Total RNA from mouse livers were extracted and HBV pC/pgRNA levels measured by qRT-PCR. Data are expressed relative to vehicle treated group (mean ± SEM, n = 2 for vehicle group and 3 for SR9009 group). f Peripheral HBeAg, HBsAg and HBV DNA levels were quantified (mean ± SEM, n = 2 for vehicle group and 3 for SR9009 group, Two way ANOVA analysis). *p < 0.05, **p< 0.01, ***p<0.001. Data are provided in the accompanying Source Data file.