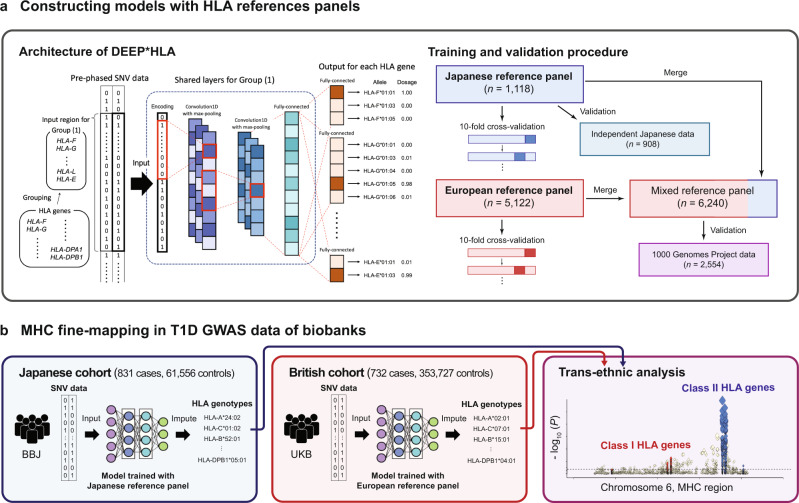
Fig. 1. An overview of the study.
a DEEP*HLA is a deep learning architecture that takes an input of pre-phased genotypes of SNVs and outputs the genotype dosages of HLA genes. To train a model and benchmark its performance, we used Japanese and European HLA reference panels respectively. We evaluated its accuracies in cross-validation with other methods. For the Japanese panel, we also evaluated its accuracy by applying the trained model to an independent Japanese HLA dataset. Further, we experimentally generated a mixed panel and validated its accuracy using 1KGv3 data. b We conducted transethnic MHC fine-mapping in T1D GWAS data. We performed HLA imputation for the Japanese cohort from BBJ and the British cohort from UKB using models specific for individual populations. We integrated the individual results of imputed genotypes and performed transethnic association analysis. SNV single nucleotide variant, HLA human leukocyte antigen, MHC major histocompatibility complex.