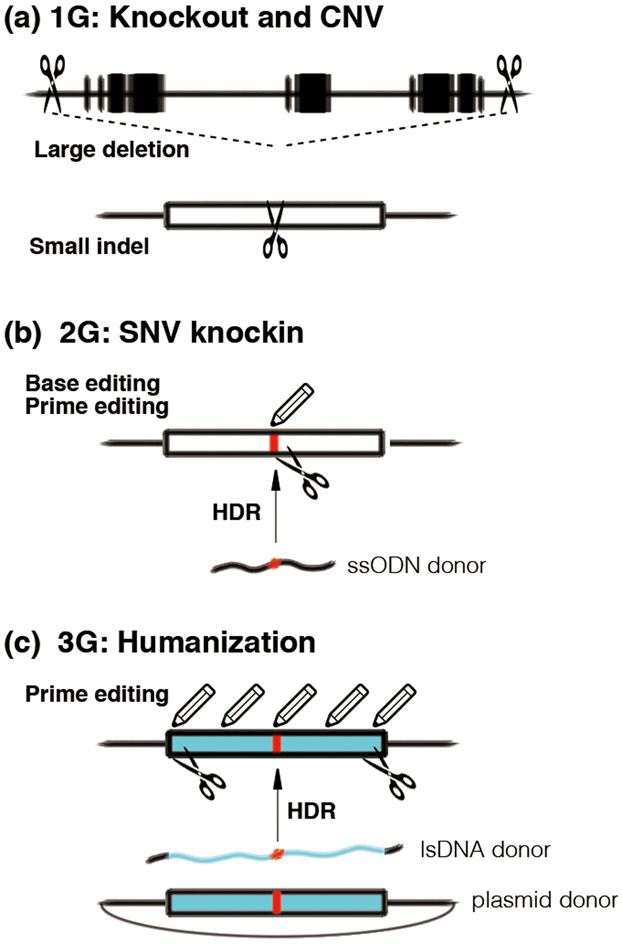
Figure 1. Genome editing technologies for next generation NHP models.
(a) 1st generation (1G) genetically modified NHP models for knockout and genomic deletion by NHEJ and large deletion. (top) large deletion of multiple genes (black boxes) by simultaneous DSBs (scissors), mimicking CNV (loss of genomic region). (bottom) Small indel of target exon by NHEJ, resulting functional gene knockout. (b) 2nd generation (2G) genetically modified NHP models for precise genome engineering and SNV knockin. SNV (red) can be precisely installed by base editing and prime editing (pencil) without DSB, or HDR with ssODN donor after DSB induction. (c) 3rd generation (3G) genetically humanized NHP models for clinical translation. Target region or exon can be humanized (blue) with SNV by prime editing without DSB or HDR with lsDNA or plasmid donor after DSB induction. NHP: non-human primate, NHEJ: non-homologous end-joining, CNV: copy number variation, SNV: single nucleotide variation, ssODN: single strand oligodeoxynucleotide, HDR: homology-directed repair, lsDNA: long single strand DNA.