Figure 1.
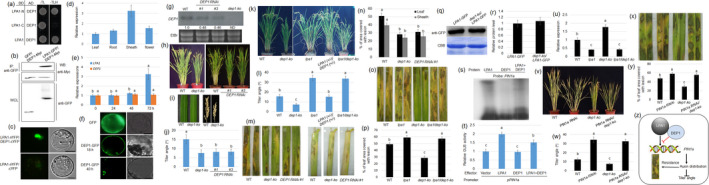
DEP1 controls the tiller angle and resistance to sheath blight disease in rice. (a) The interaction between LAP1 and DEP1 was analysed in a yeast two‐hybrid system. Interaction between activating domain (AD)‐DEP1 and binding domain (BD)‐LPA1 (full length), BD‐LPA1‐N (N‐terminal), or BD‐LAP1‐C (C‐terminal) was analysed. (b) Coimmunoprecipitation (co‐IP) was performed to analyse the interaction between DEP1 and LPA1 in tobacco leaves. DEP1‐Myc + LPA1‐GFP or DEP1‐Myc + GFP were transfected into tobacco leaves. Proteins immunoprecipitated using anti‐GFP were detected using anti‐Myc antibody. The levels of DEP1‐Myc, GFP and LPA1‐GFP from whole‐cell lysates (WCL) were detected using anti‐Myc and anti‐GFP antibodies, respectively. (c) Reconstitution of GFP fluorescence from LPA1‐nYFP + DEP1‐cCFP and LPA1‐nYFP + cCFP. Bars = 10 μm. (d) Expression patterns of DEP1 in the leaf, root, sheath and flower tissues were examined. Data indicate average ± standard error (SE) (n =3). (e) Expressions of LPA1 and DEP1 were tested after 0, 24, 48 and 72 h of R. solani AG1‐IA inoculation. Data indicate average ± standard error (SE) (n =3). (f) Free GFP and DEP1‐GFP signals were detected in the protoplasts. GFP signal was checked at 18 and 48 h. Bars = 10 μm. (g) DEP1 expression level in wild type (WT), DEP1 RNAi lines (#1 and #3) and dep1‐ko was analysed by northern blot analysis. EtBr staining was used as a loading control. The numbers shown at the bottom of the blot indicate the relative density of each band. ND: not detected. (h) Shown are 3‐month‐old WT, dep1‐ko and DEP1 RNAi lines (#1 and #3) plants. (i) The leaf and panicle of WT and dep1‐ko were photographed. (j) Tiller angles of plants from panel (h) are shown. Data indicate average ± SE (n> 10). (k) Shown are 3‐month‐old WT, dep1‐ko, lpa1, DEP1 (+/‐)/LPA1 (+/‐) and lpa1/dep1‐ko. (l) Tiller angles of the lines from panel (k) are calculated. Data indicate average ± SE (n > 10). (m) Leaves and sheath from the WT, dep1‐ko and DEP1 RNAi #1 had been inoculated with R. solani AG1‐IA. Each experiment was performed in triplicate. (n) The percentage of lesions in the leaves and sheath shown in (m) was examined. Data indicate average ± SE (n > 10). (o) Leaves from the WT, lpa1, dep1‐ko and lpa1/dep1‐ko plants were inoculated with R. solani AG1‐IA. (p) The percentage of lesions in the leaves from panel (o) was examined. Data indicate average ± SE (n > 10). (q) LPA1‐GFP protein levels in LPA1‐GFP and dep1‐ko/LPA1‐GFP transgenic plants were examined. The proteins stained with Coomassie brilliant blue (CBB) were used as the loading control. (r) The band density shown in panel (q) was calculated. Data indicate average ± SE (n =3). (s) An electrophoretic mobility shift assay (EMSA) was conducted to evaluate the affinities of LPA1 and DEP1 to the PIN1a promoter. (t) Transient expression of p35S:LPA1 alone or p35S:LPA1 and p35S:DEP1 together with a construct including the GUS gene under the control of 1.5 kb PIN1a promoter in protoplast cells. A luciferase gene driven by the 35S promoter was used as an internal control to normalize GUS expression. Error bars represent ± SE (n = 6). (u) Expression level of PIN1a in WT, lpa1, dep1‐ko and lpa1/dep1‐ko plants. Data indicate average ± SE (n =3). (w) Shown are 3‐month‐old WT, PIN1a RNAi, dep1‐ko and PIN1a RNAi/dep1‐ko plants. (v) Tiller angles of the lines from panel (u). Data indicate average ± SE (n > 10). (x) Leaves from the WT, PIN1a RNAi, dep1‐ko and PIN1a RNAi/dep1‐ko plants were inoculated with R. solani AG1‐IA and were photographed after infection. (y) The percentage of lesions in the leaves shown in panel (x) was examined. Data indicate average ± SE (n > 10). (z) Schematic diagram showing LAP1‐DEP1 regulating PIN1a transcription, and its mediated auxin distribution on sheath blight resistance and tiller angle. Different letters above the columns indicate a statistically significant difference between groups. The lesion areas in the leaves and sheath were calculated after 3 days following inoculation.
