Figure 3.
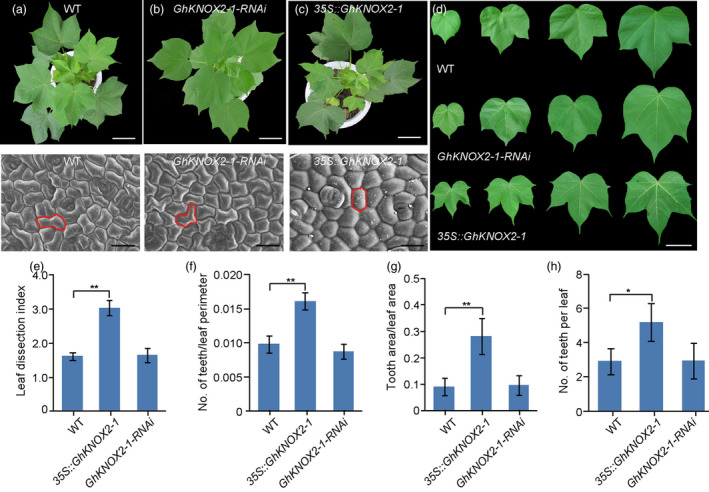
GhKNOX2‐1 regulates leaf shape in cotton. (a)–(d) Top, from left to right, phenotypic analysis of 48‐d‐old wild‐type Gossypium hirsutum (a), GhKNOX2‐1 RNAi (b), and 35S::GhKNOX2‐1 transgenic G. hirsutum plants (c). Bars = 4 cm. Bottom, from left to right, the scanning electron micrographs of the leaf epidermal cells at the base of the abaxial side of mature leaves of wild‐type, GhKNOX2‐1 RNAi and 35S::GhKNOX2‐1 transgenic plants. Bars = 50 µm. (d) Close‐up images of the leaves of 48‐d‐old wild‐type plant (top), GhKNOX2‐1 RNAi (middle) and 35S::GhKNOX2‐1 transgenic plants (bottom). Bars = 4 cm. (e)–(g) Quantitative comparisons of leaf shapes of wild‐type, GhKNOX2‐1 RNAi and 35S::GhKNOX2‐1 transgenic lines based on the leaf dissection index (perimeter2/4π × leaf area) (e), the number of teeth/ leaf perimeter (f) and the tooth area/leaf area (g). A total of 10 leaves from each line were used for each measurement. (h) Statistical analysis of number of teeth per leaf from the wild‐type, GhKNOX2‐1 RNAi and 35S::GhKNOX2‐1 transgenic plants. A total of 10 leaves from each line were used for ach measurement. Data are presented as mean ± SE. Statistical significance was determined using one‐way analysis of variance combined with Tukey's test. *P < 0.05; **P < 0.01. WT, wild type.
