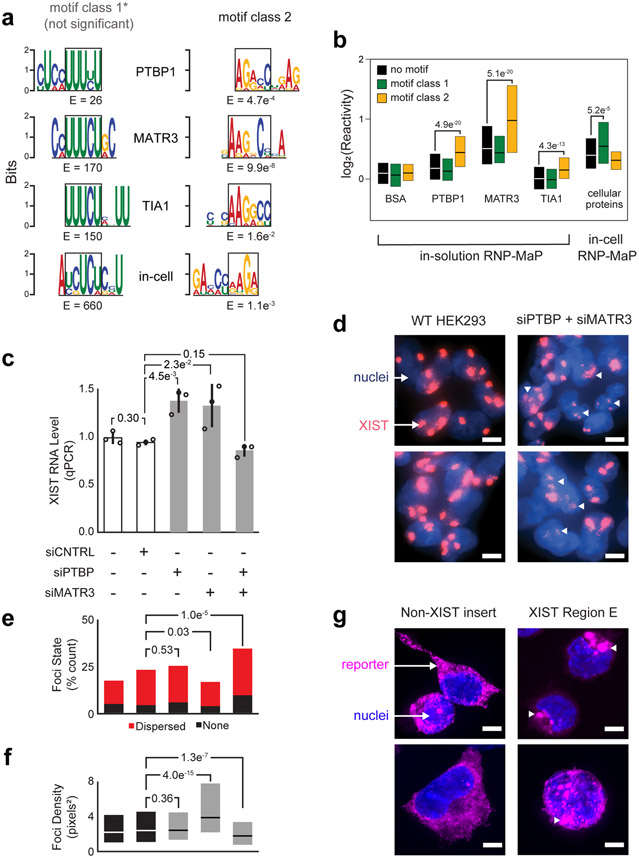
Figure 6. PTBP1 and MATR3 interactions with E region, and functional control of XIST particle formation.
(a) RNP-MaP enriched 9-mer sequence motifs (by MEME)69 using purified components for each indicated protein, and comparison with motifs observed in cell. Motifs are shown as position weighted matrices and E-values, aligned to a shared core sequence (black boxes) for class 1 and class 2 motifs. (b) Boxplots of RNP-MaP log2(reactivity) at class 1 and 2 motifs or other nucleotides (no motif) in the XIST E region, either for simplified conditions with synthetic RNA and indicated recombinant protein (in-solution) or with native XIST and cellular proteins in HEK293 cells (in-cell). The number of nucleotide reactivities included in each distribution (n), from left to right, were 631, 230, 293, 651, 235, 293, 651, 235, 293, 651, 235, 292, 668, 229, and 293. Box limits indicate first and third quartiles; central lines indicate medians. Significant increases in reactivity compared to no motif nucleotides are indicated, and – with the exception of TIA1 no motif and class 1 motif sites – reactivities in-cell and in-solution are all significantly increased compared to BSA (p <0.05, Kolmogorov-Smirnov test, one-sided). (c) Relative expression levels of XIST RNA in HEK293 cells, as a function of PTBP and MATR3 knockdown by short interfering (si)RNAs. Standard deviation (error bars), means (bar heights) of replicate measurements (n = 3, open circles), and P-values (Student’s t-test, two-sided) are shown. (d) Visualization of XIST foci for wildtype (WT) HEK293 cells and cells depleted of PTBP and MATR3 by siRNA treatment. Dispersed and non-punctate XIST foci are emphasized with white triangles. XIST in cells imaged by fluorescent in situ hybridization (FISH, red) using labeled antisense oligonucleotides; nuclei labeled with DAPI (blue). (e) Quantification of dispersed and absent XIST foci as a function of PTBP and MATR3 depletion (HEK293 cells). Conditions are ordered as per panel c. P-values (Chi-square goodness of fit test) are shown. The total number of cells counted (n) were 107, 396, 200, 286, and 304, respectively. (f) Effect of PTBP and MATR3 depletion on XIST foci density. Conditions are ordered as per panel c. P-values (Kolmogorov-Smirnov test, two-sided) are shown. The total number of XIST foci observed (n) were 311, 1000, 746, 533, and 863, respectively. Box limits indicate first and third quartiles; central lines indicate medians. (g) Localization of non-XIST control and XIST E region-containing RNA reporters (by FISH, magenta); nuclei labeled by DAPI (blue). Condensed foci formed by XIST E region reporters are emphasized by white triangles. The percentage of cells with granule phenotypes were 9% (1 of 11) and 89% (8 of 9), respectively. P-values (binomial test and chi-squared goodness of fit) were significant (7.8 e−8,, 1.13 e−13). Data from e and f were combined from two biological replicate experiments (performed and imaged on different days); d and g are representative images. Scale bars in d and g are 5 μm.