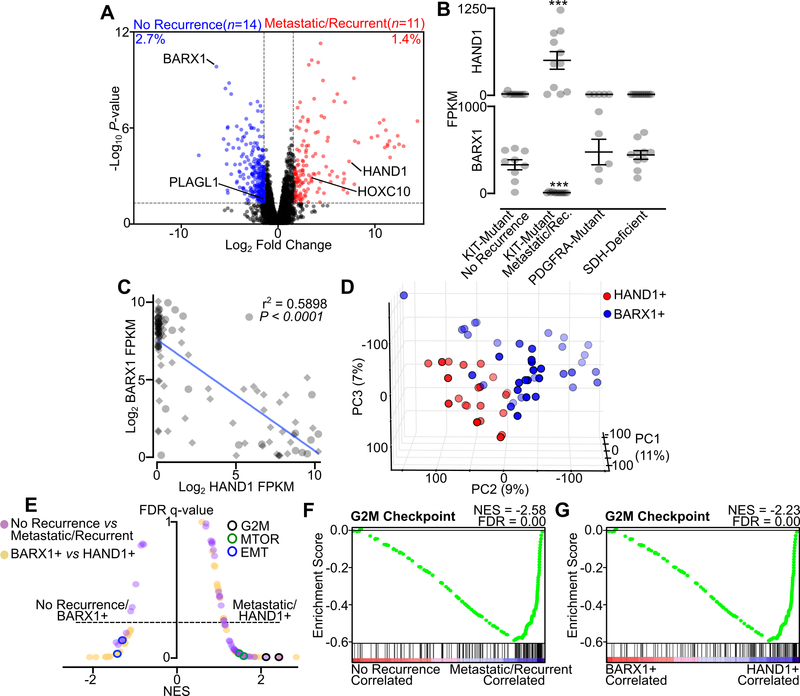
Figure 1. HAND1 is associated with metastatic GIST and a distinctive transcriptional program.
A, Volcano plot of RNA-seq data from localized and never recurrent GIST (‘No Recurrence’, n=14) or localized and later recurrent or metastatic GIST (‘Metastatic/Recurrent’, n=11). All differentially expressed TFs are labeled. The percent of genes differentially expressed is shown (n=10,000 total expressed transcripts). B, FPKM of HAND1 and BARX1 in No Recurrence KIT-mutant GIST (n=9), Metastatic/Recurrent KIT-mutant GIST (n=11), PDGFRA-mutant GIST (n=5) or SDH-deficient GIST (n=10). Data were analyzed by one-way ANOVA with Tukey’s multiple comparison test (compared to KIT-mutant localized GIST; ***,P<0.001). C, Correlation of BARX1 and HAND1 expression in all GIST samples (n=110), with the clinically annotated cohort indicated with circles and validation cohort with diamonds. The Pearson correlation is shown. D, PCA of GIST validation cohort RNA-seq (35) stratified by HAND1-positivity (n=21) and BARX1-positivity (n=39) with threshold for expression of 50 FPKM. E, Butterfly plot of all Hallmark gene sets indicating the NES and FDR q-value for the two RNA-seq data cohorts (‘No Recurrence’ and ‘Metastatic/Recurrent’ GIST in purple, BARX1- or HAND1-positive GIST in yellow). The G2M Checkpoint, MTORC1 Signaling and Epithelial Mesenchymal Transition (EMT) gene sets are indicated for each condition. F-G, GSEA showing the Hallmark G2M Checkpoint gene set in the independent GIST RNA-seq data sets.