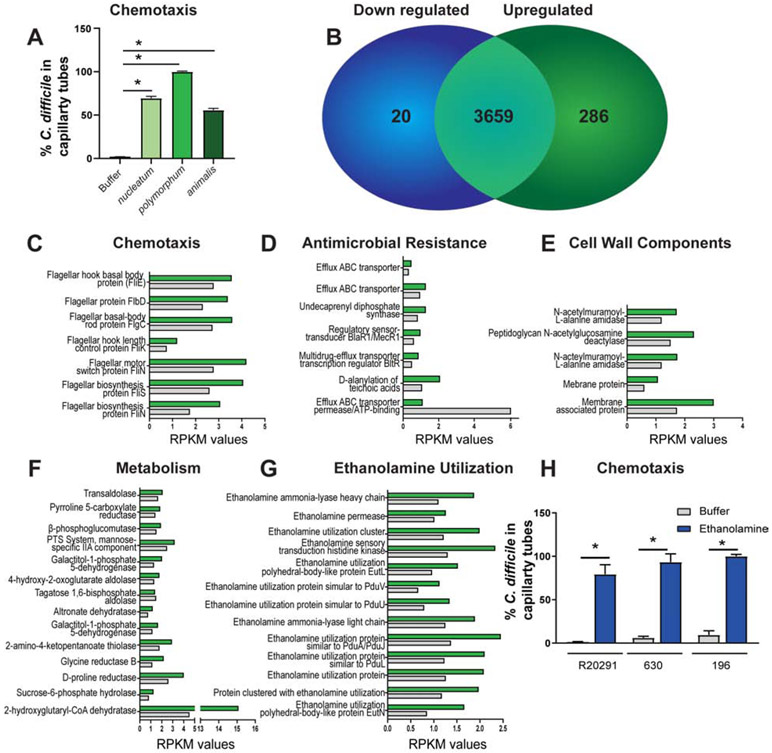
Figure 4: F. nucleatum metabolites influence C. difficile chemotaxis and gene expression.
A. Chemotaxis of fluorescently-tagged C. difficile R20291 into capillary tubes containing F. nucleatum subspecies nucleatum, polymorphum and animalis over 1 hr incubation (n=5 replicates, repeated 4 independent times). B-G. RNAseq analysis of pooled samples (n=3) of C. difficile after incubation with F. nucleatum subspecies polymorphum spent media (green bars) or C. difficile spent media (grey bars). B. 20 genes were downregulated and 286 genes were upregulated. C. difficile genes upregulated in response to F. nucleatum metabolites included (C) chemotaxis, (D) antimicrobial resistance, (E) cell wall components, (F) metabolism and (G) ethanolamine utilization. Data expressed as Reads Per Kilobase of transcript, per Million mapped reads (RPKM). H. Chemotaxis of fluorescently-tagged C. difficile strain R20291, 630 and 192 into capillary tubes containing 15 mM ethanolamine (n=5 replicates, repeated 3 independent times). One Way ANOVA, * p <0.05.