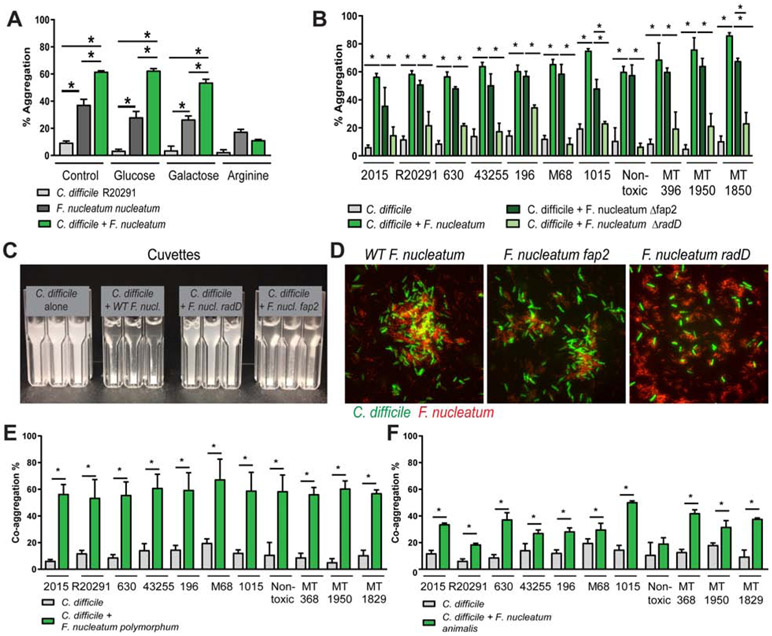
Figure 5: F. nucleatum aggregates with C. difficile via the adhesin RadD.
A. C. difficile R20291 and F. nucleatum subspecies nucleatum were incubated together anaerobically in co-aggregation buffer. Aggregation was calculated using the equation: 100%- ((OD600nm at 0 hr –OD600nn at 1 hr) x 100%). For inhibition assays, F. nucleatum subspecies nucleatum was pre-incubated for 5 min with 50 mM L-arginine, D-galactose, or D-glucose anaerobically, followed by addition of C. difficile (n=3-4 replicates, repeated 4 independent times). Significance was determined by Multi-way ANOVA. B. Aggregation was examined with 11 C. difficile strains, wild-type F. nucleatum subspecies nucleatum, as well as F. nucleatum subspecies nucleatum Δfap2 and F. nucleatum subspecies nucleatum ΔradD. C. For visualization, cuvettes were examined for aggregation in C. difficile alone, C. difficile with wild-type F. nucleatum subspecies nucleatum, Δfap2 and ΔradD. D. Microscopy of CFDA-SE tagged C. difficile R20291 (green) and F. nucleatum subspecies nucleatum (wild-type (WT), Δfap2 or ΔradD) (red) (scale bar =50 μm) (n=3). Co-aggregation was also examined with C. difficile strains and (E) F. nucleatum subspecies polymorphum and (F) F. nucleatum subspecies animalis (n=4 replicates, repeated 3 independent times). One Way ANOVA, * p <0.05.