Figure 3. Mutation of a repressive HES/ets-1 motif in the Nodal promoter yields increased Nodal transcription.
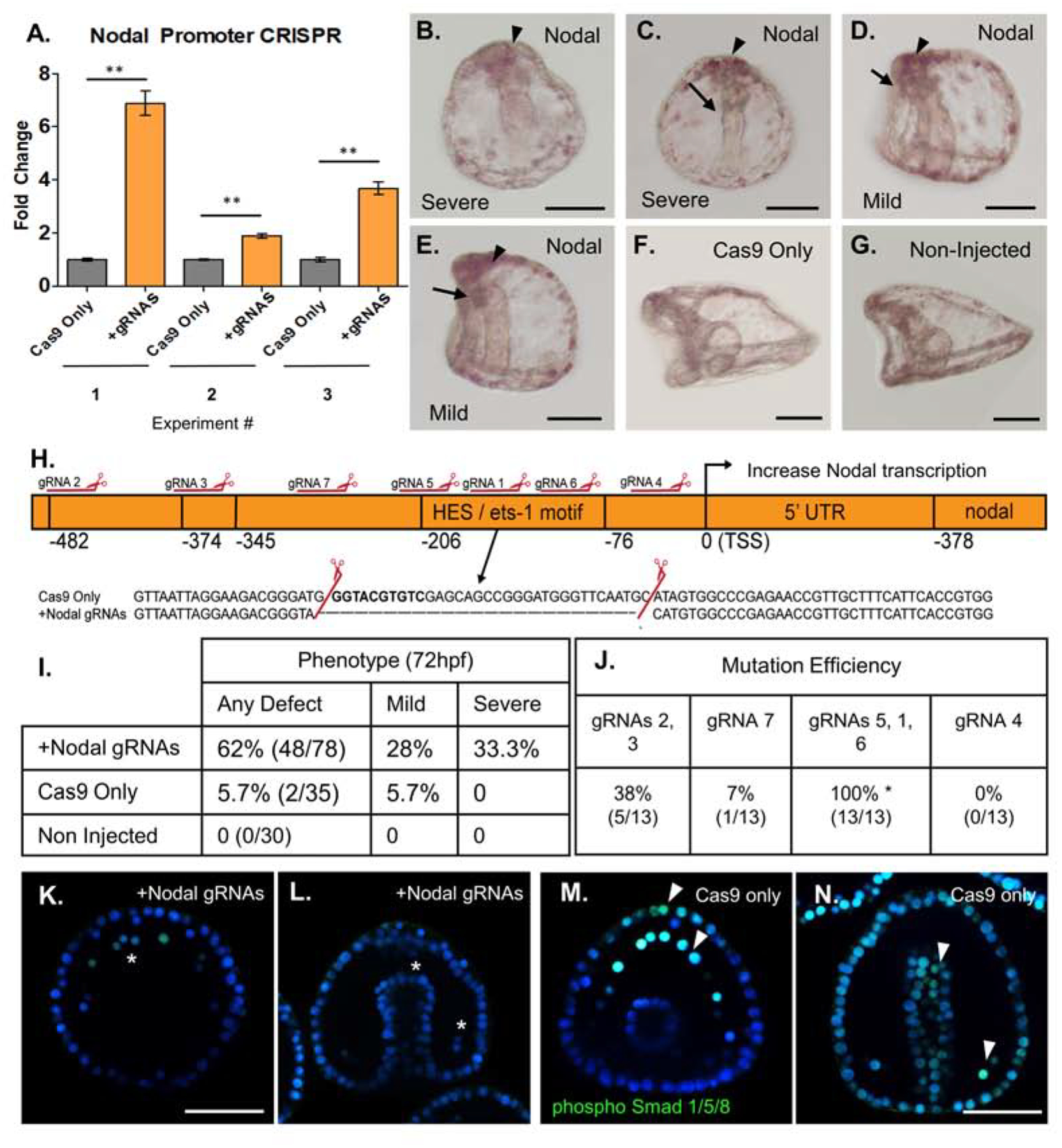
Cas9 mRNA mixed with gRNAs targeting the Nodal promoter were injected into zygotes to test sites important for Nodal expression. Cas9 mRNA-only served as an injection control and results shown are embryos from three separate matings.
A: qPCR (n=50 embryos per experiment) from each of three independent experiments demonstrated a significant (2- to 7- fold) increase in Nodal mRNA relative to Cas9 only injected control groups at 18hpf. Nodal expression was normalized to ubiquitin mRNA.
B–G: Phenotypic analysis of Nodal promoter KO embryos at 72hpf (scale bar = 60uM).
B: Severe Nodal promoter KO phenotype, formation of mouth not observed, absent dorsal-ventral flattening.. Note apical clustering of pigment cells (arrowhead).
C: Severe polarity defect at 72hpf. Gut is straight along Dorsal-Ventral axis, with no formation of mouth (arrow). Note apical clustering of pigment cells (arrowhead).
D. Mild phenotype Nodal promoter mutant embryo has normal flattening of oral and anal surfaces of the embryo, but formation of mouth is defective or absent (arrow). Apical clustering of pigment cells observed (arrowhead).
E: Mild phenotype, L/R polarity is intact, but gut formation is defective (arrow).
F: Cas9-only injected control embryo 72hpf (lateral view) shows normal gut development and flattening of oral surface and extended aboral ectoderm.
G: Age-matched control embryo 72hpf (lateral view) with normal oral-aboral patterning, gut structures, and development.
H. Nodal promoter map with mutation and repressive HES/ets-1 motif highlighted. Mutations identified through single embryo PCR genotyping demonstrated a mutation within this region, corresponding to cleavage by gRNAs 5,1 and 6. The causal mutation corresponding to the observed 2–8 fold Nodal overexpression is highlighted in the inset.
I. Summary of Nodal promoter mutant phenotypes at 72hpf. All defects are summarized in the left column, with a breakdown of mild (gut defect, oral-aboral rounding) and severe (three or more body plan defects) phenotypes in two columns.
J. Summary of gRNA mutation efficiencies at each site, as quantified from single embryo genotyping. gRNAs are grouped by their location within the promoter region. Some gRNAs produce no mutations, while others generate rather large deletions when active in pairs or a group of three. The causal deletion of the repressor HES/ets-1 site was produced by gRNAs1, 5, and 6 with 100% efficiency in sequenced individual embryos with a severe phenotype.
K. Nodal overexpressing embryo with nuclear localization of phosphorylated Smad 1/5/8 (green). pSmad 1/5/8 marks BMP2/4 responsive cells, including PMCs (asterisks). Nodal signaling is inhibitory to BMP 2/4 signaling, thus in Nodal overexpression mutants, only five pSmad1/5/8+ cells are visible in the PMC ring at 48hpf. (Scale bar = 60uM).
L. Nodal overexpressing embryos viewed dorsally demonstrate defective gut elongation, as well as an absence of pSmad 1/5/8+ nuclear signal. pSmad 1/5/8 is normally observed in response to BMP2/4, but is diminished in Nodal overexpressing mutants at 48hpf. (wildtype PMCs shown with arrowhead).
M. Phosphorylated Smad 1/5/8 nuclear signal in a Cas9 only injected embryo, showing thirteen PMCs with strong pSmad 1/5/8+ nuclear signal, and significant signal seen also in the anterior-most ectoderm (arrowheads).
N. Cas9 only control embryos viewed dorsally showing normal gut development at 48hpf, as well as phosphorylated Smad 1/5/8 nuclear signal in the BMP2/4 responsive cells (PMCs shown with arrowhead, and broadly in the ectoderm).
