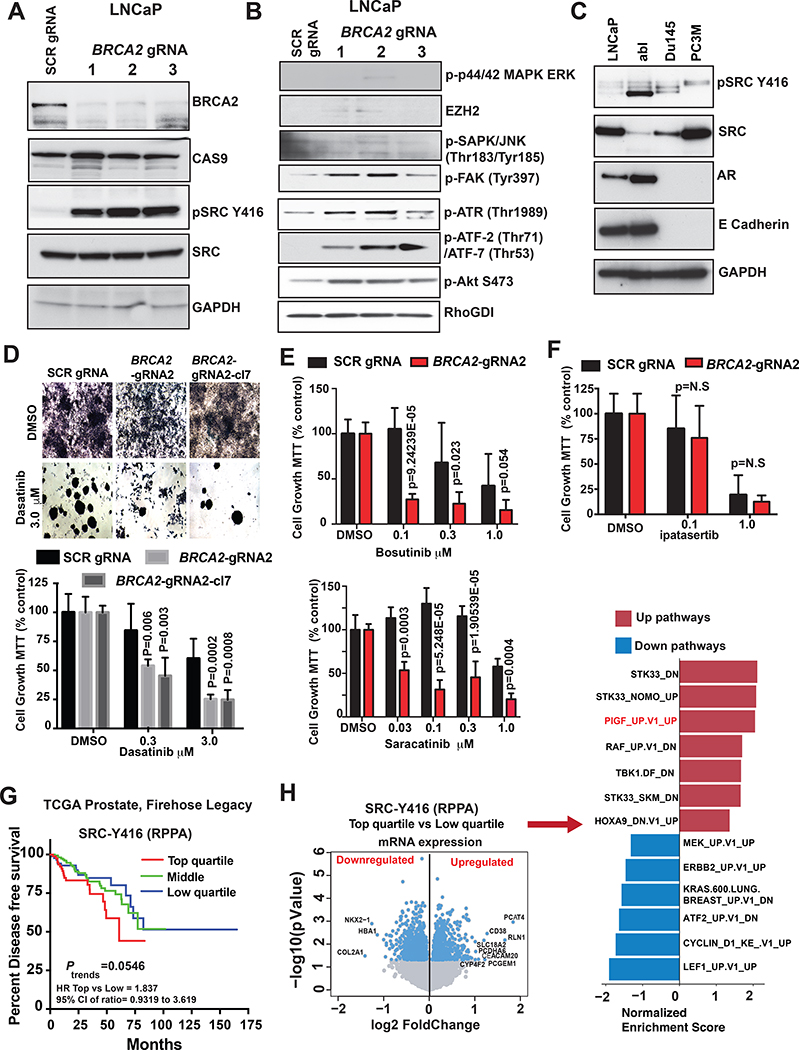
Figure 2. BRCA2 deletion induces SRC kinase activation and increases sensitivity of prostate cancer cells to dasatinib.
(A&B) LNCaP cells were transduced with three different guide RNAs (gRNAs) targeting BRCA2. Cells infected with scrambled (SCR) gRNA were used as control. CAS9, total SRC, GAPDH and RhoGDI served as loading controls. (C) Levels of phosphorylated SRC at tyrosine 416 (pSRC Y416), total SRC, androgen receptor (AR), and E-cadherin were assessed by western blot in prostate cancer cell lines. GAPDH was used as loading control. (D) Cells were treated with 0.3 and 3.0 μM dasatinib for 7 days. The equivalent volume of DMSO was used as placebo treatment. After 7 days cells were treated with 0.5 mg/mL MTT and micrographed in 40x magnification (top). The bar graph shows the changes in cell growth percentage compared to scrambled (SCR) gRNA and DMSO treated samples (bottom). P values were calculated by Student t-test. (E) LNCaP SCR control and BRCA2-gRNA 2 cells were counted and plated in 96-well plates (2500 cells/well in 100 μL media). Cells were treated with Indicated concentration of bosutinib (top) or saracatinib (bottom) for 7 days. The equivalent volume of DMSO was used as a placebo treatment. Cells were treated with 0.5 mg/mL MTT and the graphs represent cell viability (MTT count, % control). P values were calculated by Student t-test. (F) Indicated cells (LNCaP SCR control and BRCA2-gRNA 2) were counted and plated in 96-well plates (2500 cells/well in 100 μL media) and treated with 0.1 and 1.0 μM concentration of ipatasertib for 7 days. The equivalent volume of DMSO was used as a placebo treatment. Cells were treated with 0.5 mg/mL MTT, measured in a plate reader at 570 nM and represented in the form of a bar graph. P values were calculated by Student t-test. (G) Samples from patients in The Cancer Genome Atlas (TCGA) Firehose Legacy cohort were divided into 4 quartiles based on levels of phospho-SRC at Y416, reverse-phase protein arrays (RPPA). Kaplan-Meier curves were used to compare disease-free survival. Log-rank test was performed to examine significance. (H) Volcano plot shows genes that are altered in phospho-SRC Y416 high (upper quartile RPPA value) cases compared to samples with phospho-SRC Y416 low (lower quartile RPPA value). The bar graph to the right of the volcano plot shows the oncogenic pathways that are altered in phospho-SRC Y416 high vs low localized PC tumors of The Cancer Genome Atlas (TCGA) Firehose Legacy cohort. Pathway analyses were performed using GSEA (c6 oncogenic signature). The altered (based on normalized enrichment score [NES]) pathways are shown.