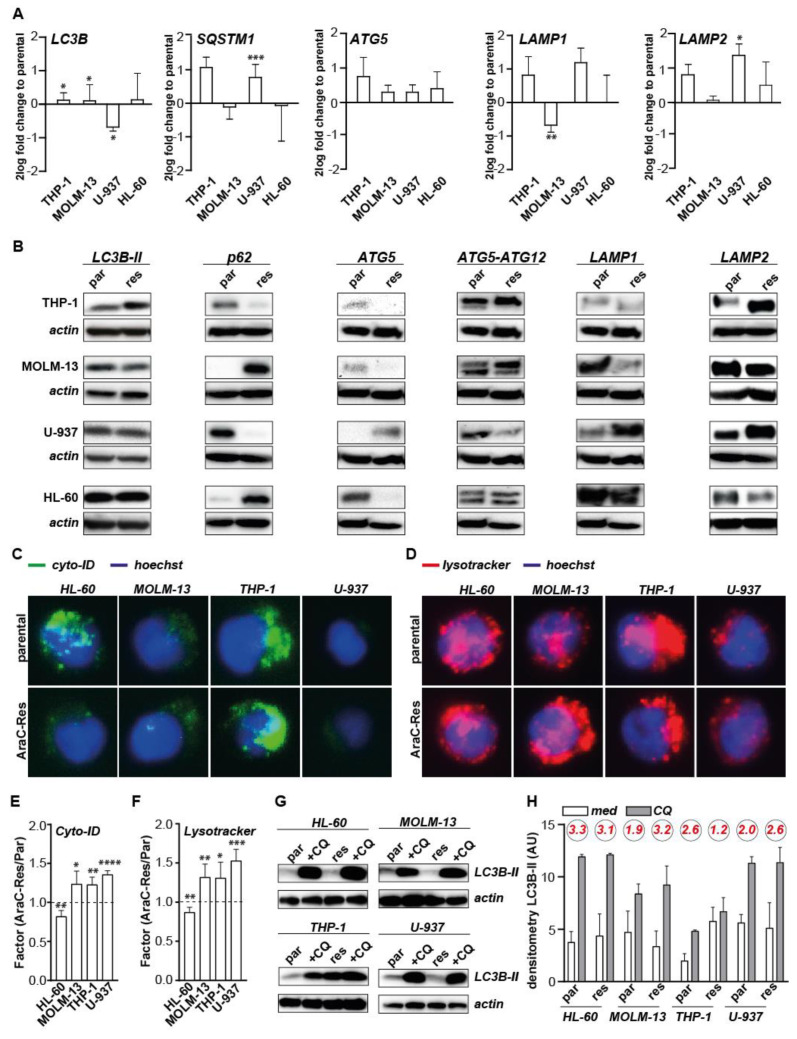
Figure 2.
Expression of general autophagy genes and proteins is not elevated in AraC resistant AML cells. (A) mRNA expression levels of autophagy genes measured in all cell line pairs and presented as a log fold change of the AraC-Res cell line compared to the parental cell line (n = 3). (B) Representative protein expression levels of autophagy-related proteins (LC3B-II (14 kDa), p62 (62 kDa), ATG5 (32 kDa), ATG5-ATG12 (55 kDa), LAMP1 (130 kDa), LAMP2 (130 kDa), and loading control beta-actin (42 kDa) in parental and AraC-Res cell lines as determined by Western blot (n = 3). (C) Representative fluorescent pictures of parental and AraC-Res cell lines stained for basal autophagosomal content using Cyto-ID, captured at 40× magnification. (D) As in (C) but stained for basal lysosomal content using lysotracker. (E,F) The difference in basal cyto-ID and lysotracker signal between AraC-Res and parental cell lines (depicted as a factor, AraC-Res/parental) as measured using flow cytometry (n = 4). (G) Representative Western blots showing the basal autophagic flux in parental versus AraC-Res cell lines using chloroquine (CQ, 50 μM for 6 h) (n = 2). (H) Densitometry measurements of the experiment as shown in (G), showing the factor increase in LC3B-II in CQ treated versus untreated cells in red (n = 2). Significance was tested using a student’s t-test. p values are indicated as: **** p < 0.0001, *** p < 0.001, ** p < 0.01, and * p < 0.05.